Linc01234 promotes cell proliferation and metastasis in oral squamous cell carcinoma via miR-433/PAK4 axis
- PMID: 32041570
- PMCID: PMC7011552
- DOI: 10.1186/s12885-020-6541-0
Linc01234 promotes cell proliferation and metastasis in oral squamous cell carcinoma via miR-433/PAK4 axis
Abstract
Background: Increasing studies have demonstrated that long non-coding RNAs (lncRNAs) play an important role in tumor progression. However, the potential biological functions and clinical importance of Linc01234 in oral squamous cell carcinoma (OSCC) remain unclear.
Methods: We evaluated the expression profile and prognostic value of Linc01234 in OSCC tissues by RT-qPCR. Then, functional in vitro experiments were performed to investigate the effects of Linc01234 on tumor growth, migration and invasion in OSCC. Mechanistically, RT-qPCR, bioinformatic analysis and dual luciferase reporter assays were performed to identify a competitive endogenous RNA (ceRNA) mechanism involving Linc01234, miR-433-3p and PAK4.
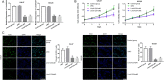
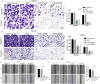
Results: We found that Linc01234 was clearly upregulated in OSCC tissues and cell lines, and its level was positively associated with T stage, lymph node metastasis, differentiation and poor prognosis of patients with OSCC. Our results shown that Linc01234 inhibited cell proliferation and metastatic abilities in CAL27 and SCC25 cells following its knockdown. Mechanistic analysis indicated that Linc01234 may act as a ceRNA (competing endogenous RNA) of miR-433-3p to relieve the repressive effect of miR-433-3p on its target PAK4.
Conclusions: Our results indicated that Linc01234 promotes OSCC progression through the Linc01234/miR-433/PAK4 axis and might be a potential therapeutic target for OSCC.
Keywords: Linc01234; OSCC; PAK4; ceRNA; miR-433.
Conflict of interest statement
The Authors declare no conflicts of interest.
Figures



Similar articles
-
Long non-coding RNA CCAT1 is a prognostic biomarker for the progression of oral squamous cell carcinoma via miR-181a-mediated Wnt/β-catenin signaling pathway.Cell Cycle. 2019 Nov;18(21):2902-2913. doi: 10.1080/15384101.2019.1662257. Epub 2019 Sep 4. Cell Cycle. 2019. PMID: 31599709 Free PMC article.
-
Long noncoding RNA SNHG20 regulates cell migration, invasion, and proliferation via the microRNA-19b-3p/RAB14 axis in oral squamous cell carcinoma.Bioengineered. 2021 Dec;12(1):3993-4003. doi: 10.1080/21655979.2021.1950278. Bioengineered. 2021. PMID: 34282711 Free PMC article.
-
Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR‑138 and releasing EZH2 in oral squamous cell carcinoma.Int J Oncol. 2018 Mar;52(3):901-912. doi: 10.3892/ijo.2018.4247. Epub 2018 Jan 16. Int J Oncol. 2018. PMID: 29344674
-
Novel insights on oral squamous cell carcinoma management using long non-coding RNAs.Oncol Res. 2024 Sep 18;32(10):1589-1612. doi: 10.32604/or.2024.052120. eCollection 2024. Oncol Res. 2024. PMID: 39308526 Free PMC article. Review.
-
Exosomal Biomarkers for Prognosis in Oral Squamous Cell Carcinoma-A Systematic Review of Emerging Technologies.J Craniofac Surg. 2025 Jun 1;36(4):1418-1424. doi: 10.1097/SCS.0000000000011104. Epub 2025 Feb 26. J Craniofac Surg. 2025. PMID: 40009435
Cited by
-
Non-Coding RNAs in Oral Cancer: Emerging Roles and Clinical Applications.Cancers (Basel). 2023 Jul 25;15(15):3752. doi: 10.3390/cancers15153752. Cancers (Basel). 2023. PMID: 37568568 Free PMC article. Review.
-
N6-methyladenosine-related lncRNAs is a potential marker for predicting prognosis and immunotherapy in ovarian cancer.Hereditas. 2022 Mar 18;159(1):17. doi: 10.1186/s41065-022-00222-3. Hereditas. 2022. PMID: 35303965 Free PMC article.
-
The role of p21-activated kinase 4 in the progression of oral squamous cell carcinoma by targeting PI3K-AKT signaling pathway.Clin Transl Oncol. 2023 Mar;25(3):739-747. doi: 10.1007/s12094-022-02980-y. Epub 2023 Jan 2. Clin Transl Oncol. 2023. PMID: 36593383
-
Long Non-Coding RNA (lncRNA) in Oral Squamous Cell Carcinoma: Biological Function and Clinical Application.Cancers (Basel). 2021 Nov 26;13(23):5944. doi: 10.3390/cancers13235944. Cancers (Basel). 2021. PMID: 34885054 Free PMC article. Review.
-
LncRNA GNAS-AS1 facilitates ER+ breast cancer cells progression by promoting M2 macrophage polarization via regulating miR-433-3p/GATA3 axis.Biosci Rep. 2020 Jul 31;40(7):BSR20200626. doi: 10.1042/BSR20200626. Biosci Rep. 2020. PMID: 32538432 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

