Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy
- PMID: 32041682
- PMCID: PMC7012603
- DOI: 10.7554/eLife.52546
Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy
Abstract
In higher plants, germline differentiation occurs during a relatively short period within developing flowers. Understanding of the mechanisms that govern germline differentiation lags behind other plant developmental processes. This is largely because the germline is restricted to relatively few cells buried deep within floral tissues, which makes them difficult to study. To overcome this limitation, we have developed a methodology for live imaging of the germ cell lineage within floral organs of Arabidopsis using light sheet fluorescence microscopy. We have established reporter lines, cultivation conditions, and imaging protocols for high-resolution microscopy of developing flowers continuously for up to several days. We used multiview imagining to reconstruct a three-dimensional model of a flower at subcellular resolution. We demonstrate the power of this approach by capturing male and female meiosis, asymmetric pollen division, movement of meiotic chromosomes, and unusual restitution mitosis in tapetum cells. This method will enable new avenues of research into plant sexual reproduction.
Keywords: A. thaliana; SPIM; cell biology; flower; germline; light sheet microscopy; live cell imaging; meiosis; plant biology.
© 2020, Valuchova et al.
Conflict of interest statement
SV, PM, JP, JK, MK, PB, SH, PT, KR No competing interests declared
Figures
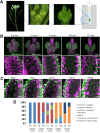
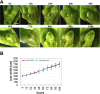
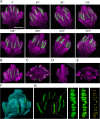
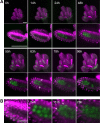

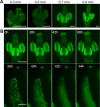
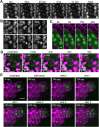
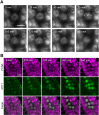


Comment in
-
Looking below the surface in plants.Elife. 2020 Feb 11;9:e54984. doi: 10.7554/eLife.54984. Elife. 2020. PMID: 32043974 Free PMC article.
Similar articles
-
Germline specification and function in plants.Annu Rev Plant Biol. 2011;62:461-84. doi: 10.1146/annurev-arplant-042110-103824. Annu Rev Plant Biol. 2011. PMID: 21332359 Review.
-
Application of Chemical Inhibitors in Live Cell Imaging of Plant Meiosis Using Light Sheet Fluorescence Microscopy.Methods Mol Biol. 2022;2484:93-105. doi: 10.1007/978-1-0716-2253-7_8. Methods Mol Biol. 2022. PMID: 35461447
-
Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development.PLoS Biol. 2011 Sep;9(9):e1001155. doi: 10.1371/journal.pbio.1001155. Epub 2011 Sep 20. PLoS Biol. 2011. PMID: 21949639 Free PMC article.
-
Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis.Mol Plant. 2008 Jul;1(4):564-74. doi: 10.1093/mp/ssn033. Mol Plant. 2008. PMID: 19825562 Review.
-
High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy.Plant J. 2011 Oct;68(2):377-85. doi: 10.1111/j.1365-313X.2011.04692.x. Epub 2011 Aug 4. Plant J. 2011. PMID: 21711399
Cited by
-
OpSeF: Open Source Python Framework for Collaborative Instance Segmentation of Bioimages.Front Bioeng Biotechnol. 2020 Oct 6;8:558880. doi: 10.3389/fbioe.2020.558880. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33117778 Free PMC article.
-
Imaging flowers: a guide to current microscopy and tomography techniques to study flower development.J Exp Bot. 2020 May 30;71(10):2898-2909. doi: 10.1093/jxb/eraa094. J Exp Bot. 2020. PMID: 32383442 Free PMC article. Review.
-
OME-Zarr: a cloud-optimized bioimaging file format with international community support.Histochem Cell Biol. 2023 Sep;160(3):223-251. doi: 10.1007/s00418-023-02209-1. Epub 2023 Jul 10. Histochem Cell Biol. 2023. PMID: 37428210 Free PMC article.
-
A Protocol for Live Imaging of Arabidopsis Gynoecium Development Using Confocal Microscopy.Methods Mol Biol. 2025;2900:351-362. doi: 10.1007/978-1-0716-4398-3_23. Methods Mol Biol. 2025. PMID: 40380073
-
Live Cell Imaging of Male Meiosis in Arabidopsis by a Landmark-based System.Bio Protoc. 2020 May 5;10(9):e3611. doi: 10.21769/BioProtoc.3611. eCollection 2020 May 5. Bio Protoc. 2020. PMID: 33659575 Free PMC article.
References
-
- Armstrong SJ, Franklin FCH, Jones GH. A meiotic time-course for Arabidopsis thaliana. Sexual Plant Reproduction. 2003;16:141–149. doi: 10.1007/s00497-003-0186-4. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

