Sorting nexin 17 (SNX17) links endosomal sorting to Eps15 homology domain protein 1 (EHD1)-mediated fission machinery
- PMID: 32041776
- PMCID: PMC7086038
- DOI: 10.1074/jbc.RA119.011368
Sorting nexin 17 (SNX17) links endosomal sorting to Eps15 homology domain protein 1 (EHD1)-mediated fission machinery
Abstract
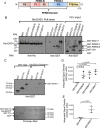
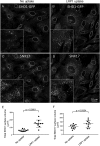
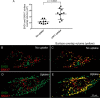
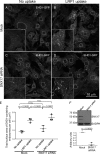
Following endocytosis, receptors that are internalized to sorting endosomes are sorted to different pathways, in part by sorting nexin (SNX) proteins. Notably, SNX17 interacts with a multitude of receptors in a sequence-specific manner to regulate their recycling. However, the mechanisms by which SNX17-labeled vesicles that contain sorted receptors bud and undergo vesicular fission from the sorting endosomes remain elusive. Recent studies suggest that a dynamin-homolog, Eps15 homology domain protein 1, catalyzes fission and releases endosome-derived vesicles for recycling to the plasma membrane. However, the mechanism by which EHD1 is coupled to various receptors and regulates their recycling remains unknown. Here we sought to characterize the mechanism by which EHD1 couples with SNX17 to regulate recycling of SNX17-interacting receptors. We hypothesized that SNX17 couples receptors to the EHD1 fission machinery in mammalian cells. Coimmunoprecipitation experiments and in vitro assays provided evidence that EHD1 and SNX17 directly interact. We also found that inducing internalization of a SNX17 cargo receptor, low-density lipoprotein receptor-related protein 1 (LRP1), led to recruitment of cytoplasmic EHD1 to endosomal membranes. Moreover, surface rendering and quantification of overlap volumes indicated that SNX17 and EHD1 partially colocalize on endosomes and that this overlap further increases upon LRP1 internalization. Additionally, SNX17-containing endosomes were larger in EHD1-depleted cells than in WT cells, suggesting that EHD1 depletion impairs SNX17-mediated endosomal fission. Our findings help clarify our current understanding of endocytic trafficking, providing significant additional insight into the process of endosomal fission and connecting the sorting and fission machineries.
Keywords: Eps15 homology domain protein 1 (EHD1); LDL receptor–related protein 1 (LRP1); endocytosis; endosomal fission; endosome; intracellular trafficking; receptor; receptor endocytosis; receptor recycling; retromer; sorting; sorting nexin (SNX); sorting nexin 17 (SNX17); trafficking; vesicles.
© 2020 Dhawan et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures

Similar articles
-
Eps15 Homology Domain Protein 4 (EHD4) is required for Eps15 Homology Domain Protein 1 (EHD1)-mediated endosomal recruitment and fission.PLoS One. 2020 Sep 23;15(9):e0239657. doi: 10.1371/journal.pone.0239657. eCollection 2020. PLoS One. 2020. PMID: 32966336 Free PMC article.
-
A sorting nexin 17-binding domain within the LRP1 cytoplasmic tail mediates receptor recycling through the basolateral sorting endosome.Traffic. 2013 Jul;14(7):823-38. doi: 10.1111/tra.12076. Epub 2013 May 8. Traffic. 2013. PMID: 23593972 Free PMC article.
-
EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval.Traffic. 2007 Dec;8(12):1873-1886. doi: 10.1111/j.1600-0854.2007.00652.x. Epub 2007 Oct 7. Traffic. 2007. PMID: 17868075
-
Towards a molecular understanding of endosomal trafficking by Retromer and Retriever.Traffic. 2019 Jul;20(7):465-478. doi: 10.1111/tra.12649. Traffic. 2019. PMID: 30993794 Review.
-
Retromer and sorting nexins in endosomal sorting.Biochem Soc Trans. 2015 Feb;43(1):33-47. doi: 10.1042/BST20140290. Biochem Soc Trans. 2015. PMID: 25619244 Review.
Cited by
-
Cellular functions and intrinsic attributes of the ATP-binding Eps15 homology domain-containing proteins.Protein Sci. 2020 Jun;29(6):1321-1330. doi: 10.1002/pro.3860. Epub 2020 Apr 11. Protein Sci. 2020. PMID: 32223019 Free PMC article. Review.
-
Neurotoxic Methamphetamine Doses Alter CDCel-1 Levels and Its Interaction with Vesicular Monoamine Transporter-2 in Rat Striatum.bioRxiv [Preprint]. 2024 Jul 23:2024.07.21.604458. doi: 10.1101/2024.07.21.604458. bioRxiv. 2024. PMID: 39091864 Free PMC article. Preprint.
-
Eps15 Homology Domain Protein 4 (EHD4) is required for Eps15 Homology Domain Protein 1 (EHD1)-mediated endosomal recruitment and fission.PLoS One. 2020 Sep 23;15(9):e0239657. doi: 10.1371/journal.pone.0239657. eCollection 2020. PLoS One. 2020. PMID: 32966336 Free PMC article.
-
Assembly and fission of tubular carriers mediating protein sorting in endosomes.Nat Rev Mol Cell Biol. 2024 Oct;25(10):765-783. doi: 10.1038/s41580-024-00746-8. Epub 2024 Jun 17. Nat Rev Mol Cell Biol. 2024. PMID: 38886588 Review.
-
FERARI and cargo adaptors coordinate cargo flow through sorting endosomes.Nat Commun. 2022 Aug 8;13(1):4620. doi: 10.1038/s41467-022-32377-y. Nat Commun. 2022. PMID: 35941155 Free PMC article.
References
-
- Clairfeuille T., Mas C., Chan A. S., Yang Z., Tello-Lafoz M., Chandra M., Widagdo J., Kerr M. C., Paul B., Mérida I., Teasdale R. D., Pavlos N. J., Anggono V., and Collins B. M. (2016) A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol. 23, 921–932 10.1038/nsmb.3290 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

