Single-Cell Analysis Reveals that the Enterococcal Sex Pheromone Response Results in Expression of Full-Length Conjugation Operon Transcripts in All Induced Cells
- PMID: 32041799
- PMCID: PMC7099139
- DOI: 10.1128/JB.00685-19
Single-Cell Analysis Reveals that the Enterococcal Sex Pheromone Response Results in Expression of Full-Length Conjugation Operon Transcripts in All Induced Cells
Abstract
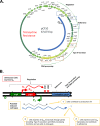
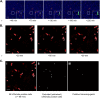
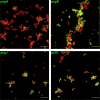
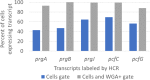
For high-frequency transfer of pCF10 between E. faecalis cells, induced expression of the pCF10 genes encoding conjugative machinery from the prgQ operon is required. This process is initiated by the cCF10 (C) inducer peptide produced by potential recipient cells. The expression timing of prgB, an "early" gene just downstream of the inducible promoter, has been studied extensively in single cells. However, several previous studies suggest that only 1 to 10% of donors induced for early prgQ gene expression actually transfer plasmids to recipients, even at a very high recipient population density. One possible explanation for this is that only a minority of pheromone-induced donors actually transcribe the entire prgQ operon. Such cells would not be able to functionally conjugate but might play another role in the group behavior of donors. Here, we sought to (i) simultaneously assess the presence of RNAs produced from the proximal (early induced transcripts [early Q]) and distal (late Q) portions of the prgQ operon in individual cells, (ii) investigate the prevalence of heterogeneity in induced transcript length, and (iii) evaluate the temporality of induced transcript expression. Using fluorescent in situ hybridization chain reaction (HCR) transcript labeling and single-cell microscopic analysis, we observed that most cells expressing early transcripts (QL, prgB, and prgA) also expressed late transcripts (prgJ, pcfC, and pcfG). These data support the conclusion that, after induction is initiated, transcription likely extends through the end of the conjugation machinery operon for most, if not all, induced cells.IMPORTANCE In Enterococcus faecalis, conjugative plasmids like pCF10 often carry antibiotic resistance genes. With antibiotic treatment, bacteria benefit from plasmid carriage; however, without antibiotic treatment, plasmid gene expression may have a fitness cost. Transfer of pCF10 is mediated by cell-to-cell signaling, which activates the expression of conjugation genes and leads to efficient plasmid transfer. Yet, not all donor cells in induced populations transfer the plasmid. We examined whether induced cells might not be able to functionally conjugate due to premature induced transcript termination. Single-cell analysis showed that most induced cells do, in fact, express all of the genes required for conjugation, suggesting that premature transcription termination within the prgQ operon does not account for failure of induced donor cell gene transfer.
Keywords: conjugation; hybridization chain reaction; plasmid transfer.
Copyright © 2020 American Society for Microbiology.
Figures


References
-
- Bandyopadhyay A. 2018. Systems analysis of pheromone signaling and antibiotic resistance transfer in Enterococcus faecalis. PhD thesis University of Minnesota, Minneapolis, MN.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

