circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway
- PMID: 32041942
- PMCID: PMC7010827
- DOI: 10.1038/s41419-020-2273-y
circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway
Abstract
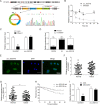
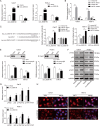
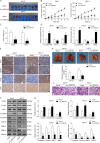
Oral squamous cell carcinoma (OSCC) is one of the most common malignancies and has a poor prognosis. Circular RNA (circRNA) has been increasingly recognized as a crucial contributor to carcinogenesis. circRNA_0000140 has been aberrantly expressed in OSCC, but its role in tumor growth and metastasis remains largely unclear. Sanger sequencing, actinomycin D, and RNase R treatments were used to confirm head-to-tail junction sequences and the stability of circ_0000140. In vitro cell activities, including proliferation, migration, invasion, and apoptosis, were determined by colony formation, transwell, and flow cytometry assays. The expression levels of circ_0000140, Hippo signaling pathway, and serial epithelial-mesenchymal transition (EMT) markers were measured by quantitative real-time PCR, western blotting, immunofluorescence, and immunohistochemistry. Dual luciferase reporter assays and Argonaute 2-RNA immunoprecipitation assays were performed to explore the interplay among circ_0000140, miR-31, and LATS2. Subcutaneous tumor growth was observed in nude mice, in which in vivo metastasis was observed following tail vein injection of OSCC cells. circ_0000140 is derived from exons 7 to 10 of the KIAA0907 gene. It was down-regulated in OSCC tissues and cell lines, and correlated negatively with poor prognostic outcomes in OSCC patients. Gain-of-function experiments demonstrated that circ_0000140 enhancement suppressed cell proliferation, migration, and invasion, and facilitated cell apoptosis in vitro. In xenograft mouse models, overexpression of circ_0000140 was able to repress tumor growth and lung metastasis. Furthermore, mechanistic studies showed that circ_0000140 could bind with miR-31 and up-regulate its target gene LATS2, thus affecting OSCC cellular EMT. Our findings demonstrated the roles of circ_0000140 in OSCC tumorigenesis as well as in metastasis, and circ_0000140 exerts its tumor-suppressing effect through miR-31/LATS2 axis of Hippo signaling pathway in OSCC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Circ-PVT1/miR-106a-5p/HK2 axis regulates cell growth, metastasis and glycolytic metabolism of oral squamous cell carcinoma.Mol Cell Biochem. 2020 Nov;474(1-2):147-158. doi: 10.1007/s11010-020-03840-5. Epub 2020 Aug 1. Mol Cell Biochem. 2020. PMID: 32737775
-
hsa_circ_0072387 Suppresses Proliferation, Metastasis, and Glycolysis of Oral Squamous Cell Carcinoma Cells by Downregulating miR-503-5p.Cancer Biother Radiopharm. 2021 Feb;36(1):84-94. doi: 10.1089/cbr.2019.3371. Epub 2020 Apr 17. Cancer Biother Radiopharm. 2021. PMID: 32302508
-
Silencing circular RNA hsa_circ_009755 promotes growth and metastasis of oral squamous cell carcinoma.Genomics. 2020 Nov;112(6):5275-5281. doi: 10.1016/j.ygeno.2020.09.035. Epub 2020 Sep 18. Genomics. 2020. PMID: 32956844
-
Metastasis and cell proliferation inhibition by microRNAs and its potential therapeutic applications in OSCC: A systematic review.Pathol Res Pract. 2024 Oct;262:155532. doi: 10.1016/j.prp.2024.155532. Epub 2024 Aug 11. Pathol Res Pract. 2024. PMID: 39142242
-
Epithelial-to-mesenchymal transition in oral squamous cell carcinoma: Challenges and opportunities.Int J Cancer. 2021 Apr 1;148(7):1548-1561. doi: 10.1002/ijc.33352. Epub 2020 Oct 29. Int J Cancer. 2021. PMID: 33091960 Review.
Cited by
-
CircRNAs: Roles in regulating head and neck squamous cell carcinoma.Front Oncol. 2022 Nov 22;12:1026073. doi: 10.3389/fonc.2022.1026073. eCollection 2022. Front Oncol. 2022. PMID: 36483049 Free PMC article. Review.
-
A therapeutical insight into the correlation between circRNAs and signaling pathways involved in cancer pathogenesis.Med Oncol. 2024 Feb 4;41(3):69. doi: 10.1007/s12032-023-02275-4. Med Oncol. 2024. PMID: 38311682 Review.
-
Circular CDC like kinase 1 suppresses cell apoptosis through miR-18b-5p/Y-box protein 2 axis in oral squamous cell carcinoma.Bioengineered. 2022 Feb;13(2):4226-4234. doi: 10.1080/21655979.2022.2027174. Bioengineered. 2022. PMID: 35156507 Free PMC article.
-
Editorial: "Non-Coding RNAs in Head and Neck Squamous Cell Carcinoma".Front Oncol. 2021 Dec 23;11:785001. doi: 10.3389/fonc.2021.785001. eCollection 2021. Front Oncol. 2021. PMID: 35004307 Free PMC article. No abstract available.
-
Characterization of Cancer Stem Cell Characteristics and Development of a Prognostic Stemness Index Cell-Related Signature in Oral Squamous Cell Carcinoma.Dis Markers. 2021 Nov 9;2021:1571421. doi: 10.1155/2021/1571421. eCollection 2021. Dis Markers. 2021. PMID: 34840626 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

