STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis
- PMID: 32041943
- PMCID: PMC7010763
- DOI: 10.1038/s41389-020-0195-2
STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis
Abstract
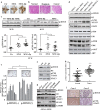
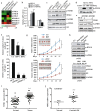
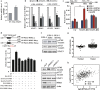
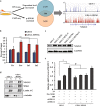
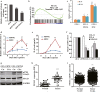
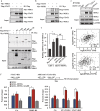
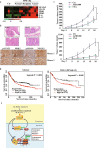
Epigenetic abnormalities contribute significantly to the development and progression of gastric cancer. However, the underlying regulatory networks from oncogenic signaling pathway to epigenetic dysregulation remain largely unclear. Here we showed that STAT3 signaling, one of the critical links between inflammation and cancer, acted as a control pathway in gastric carcinogenesis. STAT3 aberrantly transactivates the epigenetic kinase mitogen- and stress-activated protein kinase 1 (MSK1), thereby phosphorylating histone H3 serine10 (H3S10) and STAT3 itself during carcinogen-induced gastric tumorigenesis. We further identified the calcium pathway transcription factor NFATc2 as a novel downstream target of the STAT3-MSK1 positive-regulating loop. STAT3 forms a functional complex with MSK1 at the promoter of NFATc2 to promote its transcription in a H3S10 phosphorylation-dependent way, thus affecting NFATc2-related inflammatory pathways in gastric carcinogenesis. Inhibiting the STAT3/MSK1/NFATc2 signaling axis significantly suppressed gastric cancer cell proliferation and xenograft tumor growth, which provides a potential novel approach for gastric carcinogenesis intervention by regulating aberrant epigenetic and transcriptional mechanisms.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Epigenetic regulation of transcription factor promoter regions by low-dose genistein through mitogen-activated protein kinase and mitogen-and-stress activated kinase 1 nongenomic signaling.Cell Commun Signal. 2016 Aug 31;14(1):18. doi: 10.1186/s12964-016-0141-2. Cell Commun Signal. 2016. PMID: 27582276 Free PMC article.
-
GSK-3β Governs Inflammation-Induced NFATc2 Signaling Hubs to Promote Pancreatic Cancer Progression.Mol Cancer Ther. 2016 Mar;15(3):491-502. doi: 10.1158/1535-7163.MCT-15-0309. Epub 2016 Jan 28. Mol Cancer Ther. 2016. PMID: 26823495 Free PMC article.
-
p38-MAPK/MSK1-mediated overexpression of histone H3 serine 10 phosphorylation defines distance-dependent prognostic value of negative resection margin in gastric cancer.Clin Epigenetics. 2016 Aug 31;8(1):88. doi: 10.1186/s13148-016-0255-9. eCollection 2016. Clin Epigenetics. 2016. PMID: 27588146 Free PMC article.
-
Mitogen- and stress-activated protein kinase (MSK1/2) regulated gene expression in normal and disease states.Biochem Cell Biol. 2023 Jun 1;101(3):204-219. doi: 10.1139/bcb-2022-0371. Epub 2023 Feb 22. Biochem Cell Biol. 2023. PMID: 36812480 Review.
-
MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved.Sci STKE. 2003 Aug 12;2003(195):PE33. doi: 10.1126/stke.2003.195.pe33. Sci STKE. 2003. PMID: 12915720 Review.
Cited by
-
Methylomic analysis identifies C11orf87 as a novel epigenetic biomarker for GI cancers.PLoS One. 2021 Apr 22;16(4):e0250499. doi: 10.1371/journal.pone.0250499. eCollection 2021. PLoS One. 2021. PMID: 33886682 Free PMC article.
-
STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects.Biology (Basel). 2020 Jun 12;9(6):126. doi: 10.3390/biology9060126. Biology (Basel). 2020. PMID: 32545648 Free PMC article. Review.
-
Reduced DEFA5 Expression and STAT3 Activation Underlie the Submucosal Invasion of Early Gastric Cancers.Digestion. 2023;104(6):480-493. doi: 10.1159/000531790. Epub 2023 Aug 18. Digestion. 2023. PMID: 37598668 Free PMC article.
-
Short-term starvation inhibits CD36 N-glycosylation and downregulates USP7 UFMylation to alleviate RBPJ-maintained T cell exhaustion in liver cancer.Theranostics. 2025 Apr 28;15(12):5931-5952. doi: 10.7150/thno.110567. eCollection 2025. Theranostics. 2025. PMID: 40365281 Free PMC article.
-
The RNA m6A reader IGF2BP3 regulates NFAT1/IRF1 axis-mediated anti-tumor activity in gastric cancer.Cell Death Dis. 2024 Mar 6;15(3):192. doi: 10.1038/s41419-024-06566-0. Cell Death Dis. 2024. PMID: 38448411 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

