Physicochemical characterization of Pseudomonas stutzeri UFV5 and analysis of its transcriptome under heterotrophic nitrification/aerobic denitrification pathway induction condition
- PMID: 32042029
- PMCID: PMC7010759
- DOI: 10.1038/s41598-020-59279-7
Physicochemical characterization of Pseudomonas stutzeri UFV5 and analysis of its transcriptome under heterotrophic nitrification/aerobic denitrification pathway induction condition
Abstract
Biological ammonium removal via heterotrophic nitrification/aerobic denitrification (HN/AD) presents several advantages in relation to conventional removal processes, but little is known about the microorganisms and metabolic pathways involved in this process. In this study, Pseudomonas stutzeri UFV5 was isolated from an activated sludge sample from oil wastewater treatment station and its ammonium removal via HN/AD was investigated by physicochemical and molecular approaches to better understand this process and optimize the biological ammonium removal in wastewater treatment plants. Results showed that P. stutzeri UFV5 removed all the ammonium in 48-72 hours using pyruvate, acetate, citrate or sodium succinate as carbon sources, C/N ratios 6, 8, 10 and 12, 3-6% salinities, pH 7-9 and temperatures of 20-40 °C. Comparative genomics and PCR revealed that genes encoding the enzymes involved in anaerobic denitrification process are present in P. stutzeri genome, but no gene that encodes enzymes involved in autotrophic nitrification was found. Furthermore, transcriptomics showed that none of the known enzymes of autotrophic nitrification and anaerobic denitrification had their expression differentiated and an upregulation of the biosynthesis machinery and protein translation was observed, besides several genes with unknown function, indicating a non-conventional mechanism involved in HN/AD process.
Conflict of interest statement
The authors declare no competing interests.
Figures


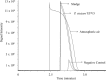
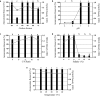
 ) and ammonium removal (■) of bacterial isolate P. stutzeri UFV5 after 72 hours of incubation in HNM medium. (a) Carbon source - the carbon sources used were sodium pyruvate (SP), sucrose (S), sodium acetate (SA), sodium citrate (SC), sodium succinate (SS) and glucose (G)), (b) pH - 3, 5, 7 and 9, (c) Carbon/Nitrogen Ratio - 4, 6, 8, 10 and 12, (d) Salinity - 0, 3, 6, 9, 12 and 15% of NaCl and (e) Temperature - 20, 25, 30, 35 and 40 °C. The graph represents the mean values of ammonium removal and optical density and their respective standard deviations. Ammonium removal means followed by at least one same letter did not differ at a 5% level of significance as determined by Tukey’s test.
) and ammonium removal (■) of bacterial isolate P. stutzeri UFV5 after 72 hours of incubation in HNM medium. (a) Carbon source - the carbon sources used were sodium pyruvate (SP), sucrose (S), sodium acetate (SA), sodium citrate (SC), sodium succinate (SS) and glucose (G)), (b) pH - 3, 5, 7 and 9, (c) Carbon/Nitrogen Ratio - 4, 6, 8, 10 and 12, (d) Salinity - 0, 3, 6, 9, 12 and 15% of NaCl and (e) Temperature - 20, 25, 30, 35 and 40 °C. The graph represents the mean values of ammonium removal and optical density and their respective standard deviations. Ammonium removal means followed by at least one same letter did not differ at a 5% level of significance as determined by Tukey’s test.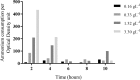


Similar articles
-
Aerobic denitrification by Pseudomonas stutzeri C3 incapable of heterotrophic nitrification.Bioprocess Biosyst Eng. 2015 Feb;38(2):407-9. doi: 10.1007/s00449-014-1271-9. Epub 2014 Aug 13. Bioprocess Biosyst Eng. 2015. PMID: 25115799
-
Heterotrophic nitrification and aerobic denitrification by the bacterium Pseudomonas stutzeri YZN-001.Bioresour Technol. 2011 Nov;102(21):9866-9. doi: 10.1016/j.biortech.2011.07.118. Epub 2011 Aug 9. Bioresour Technol. 2011. PMID: 21911288
-
Ammonium Removal by a Newly Isolated Heterotrophic Nitrification-Aerobic Denitrification Bacteria Pseudomonas Stutzeri SDU10 and Its Potential in Treatment of Piggery Wastewater.Curr Microbiol. 2020 Oct;77(10):2792-2801. doi: 10.1007/s00284-020-02085-1. Epub 2020 Jun 16. Curr Microbiol. 2020. PMID: 32556477
-
Heterotrophic nitrification and aerobic denitrification process: Promising but a long way to go in the wastewater treatment.Sci Total Environ. 2022 Jan 20;805:150212. doi: 10.1016/j.scitotenv.2021.150212. Epub 2021 Sep 10. Sci Total Environ. 2022. PMID: 34536867 Review.
-
[Advances in heterotrophic nitrification-aerobic denitrifying bacteria for nitrogen removal under extreme conditions].Sheng Wu Gong Cheng Xue Bao. 2019 Jun 25;35(6):942-955. doi: 10.13345/j.cjb.180427. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31231992 Review. Chinese.
Cited by
-
Heterotrophic nitrification and related functional gene expression characteristics of Alcaligenes faecalis SDU20 with the potential use in swine wastewater treatment.Bioprocess Biosyst Eng. 2021 Oct;44(10):2035-2050. doi: 10.1007/s00449-021-02581-z. Epub 2021 May 12. Bioprocess Biosyst Eng. 2021. PMID: 33978835
-
Pangenomic landscapes shape performances of a synthetic genetic circuit across Stutzerimonas species.mSystems. 2024 Sep 17;9(9):e0084924. doi: 10.1128/msystems.00849-24. Epub 2024 Aug 21. mSystems. 2024. PMID: 39166875 Free PMC article.
-
Network pharmacology analysis and experimental verification reveal the mechanism of the traditional Chinese medicine YU-Pingfeng San alleviating allergic rhinitis inflammatory responses.Front Plant Sci. 2022 Aug 9;13:934130. doi: 10.3389/fpls.2022.934130. eCollection 2022. Front Plant Sci. 2022. PMID: 36017263 Free PMC article.
-
Physiological adaptation and population dynamics of a nitrifying sludge exposed to ampicillin.Int Microbiol. 2024 Aug;27(4):1035-1047. doi: 10.1007/s10123-023-00452-z. Epub 2023 Nov 27. Int Microbiol. 2024. PMID: 38010565
-
Genomic insights into heterotrophic nitrifying-aerobic denitrifying bacteria from petroleum terminal effluents.Heliyon. 2024 Oct 18;10(21):e39436. doi: 10.1016/j.heliyon.2024.e39436. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39568850 Free PMC article.
References
-
- Rittmann, R. E. & McCarty, P. L. Environmental Biotechnology: Principles and Applications. McGraw-Hill, Singapore (2001).
-
- Madigan, T. M., Martinko, J. M. & Parker, J. Brock Biology of Microorganisms, Prentice Hall, 14ª edition, New York (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

