Continuous evolution of SpCas9 variants compatible with non-G PAMs
- PMID: 32042170
- PMCID: PMC7145744
- DOI: 10.1038/s41587-020-0412-8
Continuous evolution of SpCas9 variants compatible with non-G PAMs
Abstract
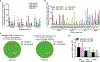
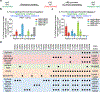
The targeting scope of Streptococcus pyogenes Cas9 (SpCas9) and its engineered variants is largely restricted to protospacer-adjacent motif (PAM) sequences containing G bases. Here we report the evolution of three new SpCas9 variants that collectively recognize NRNH PAMs (where R is A or G and H is A, C or T) using phage-assisted non-continuous evolution, three new phage-assisted continuous evolution strategies for DNA binding and a secondary selection for DNA cleavage. The targeting capabilities of these evolved variants and SpCas9-NG were characterized in HEK293T cells using a library of 11,776 genomically integrated protospacer-sgRNA pairs containing all possible NNNN PAMs. The evolved variants mediated indel formation and base editing in human cells and enabled A•T-to-G•C base editing of a sickle cell anemia mutation using a previously inaccessible CACC PAM. These new evolved SpCas9 variants, together with previously reported variants, in principle enable targeting of most NR PAM sequences and substantially reduce the fraction of genomic sites that are inaccessible by Cas9-based methods.
Conflict of interest statement
Competing Interests
The authors declare competing financial interests: S.M.M., T.W., and D.R.L. have filed patent applications on this work. D.R.L. is a consultant and co-founder of Editas Medicine, Pariwise Plants, Beam Therapeutics, and Prime Medicine, companies that use genome editing technologies.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

