Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke
- PMID: 32042176
- PMCID: PMC7523735
- DOI: 10.1038/s41593-020-0585-y
Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke
Abstract
Monocyte-derived and tissue-resident macrophages are ontogenetically distinct components of the innate immune system. Assessment of their respective functions in pathology is complicated by changes to the macrophage phenotype during inflammation. Here we find that Cxcr4-CreER enables permanent genetic labeling of hematopoietic stem cells (HSCs) and distinguishes HSC-derived monocytes from microglia and other tissue-resident macrophages. By combining Cxcr4-CreER-mediated lineage tracing with Cxcr4 inhibition or conditional Cxcr4 ablation in photothrombotic stroke, we find that Cxcr4 promotes initial monocyte infiltration and subsequent territorial restriction of monocyte-derived macrophages to infarct tissue. After transient focal ischemia, Cxcr4 deficiency reduces monocyte infiltration and blunts the expression of pattern recognition and defense response genes in monocyte-derived macrophages. This is associated with an altered microglial response and deteriorated outcomes. Thus, Cxcr4 is essential for an innate-immune-system-mediated defense response after cerebral ischemia. We further propose Cxcr4-CreER as a universal tool to study functions of HSC-derived cells.
Conflict of interest statement
Competing Interests Statement
All authors declare no competing interests.
Figures




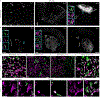


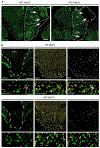

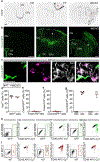

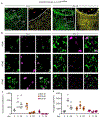




References
-
- Alliot F, Godin I & Pessac B Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res 117, 145–152 (1999). - PubMed
-
- Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

