Therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and neuroblastoma progression
- PMID: 32042322
- PMCID: PMC6993229
- DOI: 10.7150/thno.37383
Therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and neuroblastoma progression
Abstract
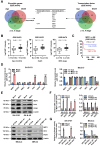
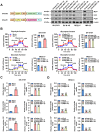
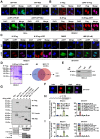
As a hallmark of metabolic reprogramming, aerobic glycolysis contributes to tumorigenesis and aggressiveness. However, the mechanisms and therapeutic strategies regulating aerobic glycolysis in neuroblastoma (NB), one of leading causes of cancer-related death in childhood, still remain elusive. Methods: Transcriptional regulators and their downstream glycolytic genes were identified by a comprehensive screening of publicly available datasets. Dual-luciferase, chromatin immunoprecipitation, real-time quantitative RT-PCR, western blot, gene over-expression or silencing, co-immunoprecipitation, mass spectrometry, peptide pull-down assay, sucrose gradient sedimentation, seahorse extracellular flux, MTT colorimetric, soft agar, matrigel invasion, and nude mice assays were undertaken to explore the biological effects and underlying mechanisms of transcriptional regulators in NB cells. Survival analysis was performed by using log-rank test and Cox regression assay. Results: Transcription factor myeloid zinc finger 1 (MZF1) was identified as an independent prognostic factor (hazard ratio=2.330, 95% confidence interval=1.021 to 3.317), and facilitated glycolysis process through increasing expression of hexokinase 2 (HK2) and phosphoglycerate kinase 1 (PGK1). Meanwhile, a 21-amino acid peptide encoded by upstream open reading frame of MZF1, termed as MZF1-uPEP, bound to zinc finger domain of Yin Yang 1 (YY1), resulting in repressed transactivation of YY1 and decreased transcription of MZF1 and downstream genes HK2 and PGK1. Administration of a cell-penetrating MZF1-uPEP or lentivirus over-expressing MZF1-uPEP inhibited the aerobic glycolysis, tumorigenesis and aggressiveness of NB cells. In clinical NB cases, low expression of MZF1-uPEP or high expression of MZF1, YY1, HK2, or PGK1 was associated with poor survival of patients. Conclusions: These results indicate that therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and NB progression.
Keywords: Yin Yang 1.; aerobic glycolysis; myeloid zinc finger 1; tumor progression; upstream open reading frame.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
HNF4A-AS1/hnRNPU/CTCF axis as a therapeutic target for aerobic glycolysis and neuroblastoma progression.J Hematol Oncol. 2020 Mar 26;13(1):24. doi: 10.1186/s13045-020-00857-7. J Hematol Oncol. 2020. Retraction in: J Hematol Oncol. 2023 Aug 15;16(1):95. doi: 10.1186/s13045-023-01493-7. PMID: 32216806 Free PMC article. Retracted.
-
YY1 drives PARP1 expression essential for PARylation of NONO in mRNA maturation during neuroblastoma progression.J Transl Med. 2024 Dec 27;22(1):1153. doi: 10.1186/s12967-024-05956-4. J Transl Med. 2024. PMID: 39731187 Free PMC article.
-
Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression.EMBO Mol Med. 2019 Dec;11(12):e10835. doi: 10.15252/emmm.201910835. Epub 2019 Nov 11. EMBO Mol Med. 2019. PMID: 31709724 Free PMC article.
-
Zinc Finger Transcription Factor MZF1-A Specific Regulator of Cancer Invasion.Cells. 2020 Jan 16;9(1):223. doi: 10.3390/cells9010223. Cells. 2020. PMID: 31963147 Free PMC article. Review.
-
The Yin and Yang of YY1 in tumor growth and suppression.Int J Cancer. 2018 Aug 1;143(3):460-465. doi: 10.1002/ijc.31255. Epub 2018 Jan 31. Int J Cancer. 2018. PMID: 29322514 Review.
Cited by
-
Integrated Bioinformatics and Experimental Approaches Identified the Role of NPPA in the Proliferation and the Malignant Behavior of Breast Cancer.J Immunol Res. 2021 Sep 27;2021:7876489. doi: 10.1155/2021/7876489. eCollection 2021. J Immunol Res. 2021. PMID: 34616853 Free PMC article.
-
SNORA37/CMTR1/ELAVL1 feedback loop drives gastric cancer progression via facilitating CD44 alternative splicing.J Exp Clin Cancer Res. 2025 Jan 16;44(1):15. doi: 10.1186/s13046-025-03278-x. J Exp Clin Cancer Res. 2025. PMID: 39815331 Free PMC article.
-
Downregulation of MEG3 promotes neuroblastoma development through FOXO1-mediated autophagy and mTOR-mediated epithelial-mesenchymal transition.Int J Biol Sci. 2020 Oct 3;16(15):3050-3061. doi: 10.7150/ijbs.48126. eCollection 2020. Int J Biol Sci. 2020. PMID: 33061817 Free PMC article.
-
Small open reading frame-encoded microproteins in cancer: identification, biological functions and clinical significance.Mol Cancer. 2025 Apr 2;24(1):105. doi: 10.1186/s12943-025-02278-x. Mol Cancer. 2025. PMID: 40170020 Free PMC article. Review.
-
Silencing of circ_0000517 suppresses proliferation, glycolysis, and glutamine decomposition of non-small cell lung cancer by modulating miR-330-5p/YY1 signal pathway.Kaohsiung J Med Sci. 2021 Dec;37(12):1027-1037. doi: 10.1002/kjm2.12440. Epub 2021 Aug 18. Kaohsiung J Med Sci. 2021. PMID: 34405943 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

