Nerve growth factor activates autophagy in Schwann cells to enhance myelin debris clearance and to expedite nerve regeneration
- PMID: 32042328
- PMCID: PMC6993217
- DOI: 10.7150/thno.40919
Nerve growth factor activates autophagy in Schwann cells to enhance myelin debris clearance and to expedite nerve regeneration
Abstract
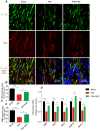
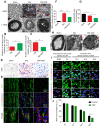
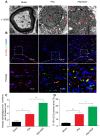
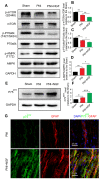
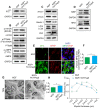
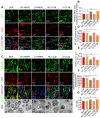
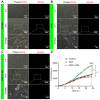
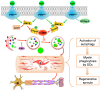
Rationale: Autophagy in Schwann cells (SCs) is crucial for myelin debris degradation and clearance following peripheral nerve injury (PNI). Nerve growth factor (NGF) plays an important role in reconstructing peripheral nerve fibers and promoting axonal regeneration. However, it remains unclear if NGF effect in enhancing nerve regeneration is mediated through autophagic clearance of myelin debris in SCs. Methods: In vivo, free NGF solution plus with/without pharmacological inhibitors were administered to a rat sciatic nerve crush injury model. In vitro, the primary Schwann cells (SCs) and its cell line were cultured in normal medium containing NGF, their capable of swallowing or clearing degenerated myelin was evaluated through supplement of homogenized myelin fractions. Results: Administration of exogenous NGF could activate autophagy in dedifferentiated SCs, accelerate myelin debris clearance and phagocytosis, as well as promote axon and myelin regeneration at early stage of PNI. These NGF effects were effectively blocked by autophagy inhibitors. In addition, inhibition of the p75 kD neurotrophin receptor (p75NTR) signal or inactivation of the AMP-activated protein kinase (AMPK) also inhibited the NGF effect as well. Conclusions: NGF effect on promoting early nerve regeneration is closely associated with its accelerating autophagic clearance of myelin debris in SCs, which probably regulated by the p75NTR/AMPK/mTOR axis. Our studies thus provide strong support that NGF may serve as a powerful pharmacological therapy for peripheral nerve injuries.
Keywords: Autophagic flux; Myelin debris clearance; Nerve growth factor (NGF); Nerve regeneration; Schwann cells.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures







References
-
- Griffin JW, Hogan MV, Chhabra AB, Deal DN. Peripheral nerve repair and reconstruction. J Bone Joint Surg Am. 2013;95:2144–51. - PubMed
-
- Forbes SJ, Rosenthal N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat Med. 2014;20:857–69. - PubMed
-
- Zhao Y, Wang Y, Gong J, Yang L, Niu C, Ni X. et al. Chitosan degradation products facilitate peripheral nerve regeneration by improving macrophage-constructed microenvironments. Biomaterials. 2017;134:64–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

