Integrated metabolomics and lipidomics profiling of hippocampus reveal metabolite biomarkers in a rat model of chronic unpredictable mild stress-induced depression
- PMID: 32042797
- PMCID: PMC6990017
- DOI: 10.21037/atm.2019.11.21
Integrated metabolomics and lipidomics profiling of hippocampus reveal metabolite biomarkers in a rat model of chronic unpredictable mild stress-induced depression
Abstract
Background: Prolonged exposure to stress triggers depression, threatening human health. Thus, to thoroughly understand the underlying pathophysiologic mechanism of chronic unpredictable mild stress (CUMS)-induced depression is urgently needed. Ultra-high-performance liquid chromatography-mass spectroscopy (UPLC-MS)-based lipidomic and metabolomic approaches has been used for discovering metabolite biomarkers to develop new diagnostic and therapeutic means. Thus, our study aimed to conduct integrated metabolomics and lipidomics to identify metabolites and lipids biomarkers in the hippocampus in rat models of CUMS-induced depression.
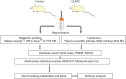
Methods: Twelve eight-week-old male Sprague-Dawley rats (weight 210±30 g) were randomly distributed to one of the following two groups (n=6): control or CUMS. Established UPLC-MS-based lipidomic and metabolomic approaches were used to determine the metabolites and lipids in the hippocampus of rats. SICMA-P and GraphPad software were performed to discover potential metabolites and lipids biomarkers in the hippocampus of rats between the two groups.
Results: A total of 35 potential metabolites and 171 lipids were identified and found to be mainly related to amino acid and lipid metabolism. These metabolites were involved in different metabolic pathways and connected to each other, which might participate in the occurrence and development of depression.
Conclusions: Our findings underlined the metabolites, lipids and metabolic pathways that were changed in the hippocampus in CUMS compared to the controls, providing novel insights in the metabolism in the hippocampus of rats and revealing the new lipid-related targets.
Keywords: Chronic unpredictable mild stress (CUMS); depression; lipidomics; metabolomics; ultra-high-performance liquid chromatography-mass spectroscopy (UPLC-MS).
2019 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Metabolomics analysis of the antidepressant prescription Danzhi Xiaoyao Powder in a rat model of Chronic Unpredictable Mild Stress (CUMS).J Ethnopharmacol. 2020 Oct 5;260:112832. doi: 10.1016/j.jep.2020.112832. Epub 2020 May 6. J Ethnopharmacol. 2020. PMID: 32387465
-
Metabolomic analysis of the hippocampus in a rat model of chronic mild unpredictable stress-induced depression based on a pathway crosstalk and network module approach.J Pharm Biomed Anal. 2021 Jan 30;193:113755. doi: 10.1016/j.jpba.2020.113755. Epub 2020 Nov 6. J Pharm Biomed Anal. 2021. PMID: 33190083
-
Metabolomics Analysis of the Prefrontal Cortex in a Rat Chronic Unpredictable Mild Stress Model of Depression.Front Psychiatry. 2022 Mar 15;13:815211. doi: 10.3389/fpsyt.2022.815211. eCollection 2022. Front Psychiatry. 2022. PMID: 35370823 Free PMC article.
-
Possibilities of Liquid Chromatography Mass Spectrometry (LC-MS)-Based Metabolomics and Lipidomics in the Authentication of Meat Products: A Mini Review.Food Sci Anim Resour. 2022 Sep;42(5):744-761. doi: 10.5851/kosfa.2022.e37. Epub 2022 Sep 1. Food Sci Anim Resour. 2022. PMID: 36133639 Free PMC article. Review.
-
Lipid Metabolite Biomarkers in Cardiovascular Disease: Discovery and Biomechanism Translation from Human Studies.Metabolites. 2021 Sep 14;11(9):621. doi: 10.3390/metabo11090621. Metabolites. 2021. PMID: 34564437 Free PMC article. Review.
Cited by
-
Exercise Alleviates Fluoride-Induced Learning and Memory Impairment in Mice: Role of miR-206-3p and PREG.Biol Trace Elem Res. 2024 Nov;202(11):5126-5144. doi: 10.1007/s12011-024-04068-w. Epub 2024 Jan 20. Biol Trace Elem Res. 2024. PMID: 38244175
-
MicroRNA-367-3p suppresses sevoflurane-induced adult rat astrocyte apoptosis by targeting BCL2L11.Exp Ther Med. 2022 Jan;23(1):9. doi: 10.3892/etm.2021.10931. Epub 2021 Oct 28. Exp Ther Med. 2022. PMID: 34815761 Free PMC article.
-
Alteration of Glycerophospholipid Metabolism in Hippocampus of Post-stroke Depression Rats.Neurochem Res. 2022 Jul;47(7):2052-2063. doi: 10.1007/s11064-022-03596-y. Epub 2022 Apr 25. Neurochem Res. 2022. PMID: 35469367
-
Study on the Effect of Different Iodine Intake on Hippocampal Metabolism in Offspring Rats.Biol Trace Elem Res. 2022 Oct;200(10):4385-4394. doi: 10.1007/s12011-021-03032-2. Epub 2021 Dec 2. Biol Trace Elem Res. 2022. PMID: 34855145
-
CD36 deficiency affects depressive-like behaviors possibly by modifying gut microbiota and the inflammasome pathway in mice.Transl Psychiatry. 2021 Jan 5;11(1):16. doi: 10.1038/s41398-020-01130-8. Transl Psychiatry. 2021. PMID: 33414380 Free PMC article.
References
LinkOut - more resources
Full Text Sources
