The analysis of pharmacokinetic and pharmacogenomic impact on gefitinib efficacy in advanced non-small cell lung cancer patients: results from a prospective cohort study
- PMID: 32042822
- PMCID: PMC6989872
- DOI: 10.21037/atm.2019.12.60
The analysis of pharmacokinetic and pharmacogenomic impact on gefitinib efficacy in advanced non-small cell lung cancer patients: results from a prospective cohort study
Abstract
Background: The current study is aimed to examine the impact of pharmacokinetics and gene polymorphisms of enzymes involving in absorption, distribution, metabolism and excretion (ADME) on the efficacy of gefitinib in non-small cell lung cancer (NSCLC) patients.
Methods: Eligible patients with indication of gefitinib treatment were prospectively enrolled in this study. Two peripheral blood samples at baseline and before cycle 2 day 1 were collected for the detection of single nucleotide polymorphisms (SNPs) of drug ADME enzymes and trough drug concentration (Ctrough) at steady state. Thirteen SNPs were genotyped using the Sequenom Massarray system. Ctrough was determined by validated high-performance liquid chromatographic method with tandem mass spectrometric (LC-MS/MS).
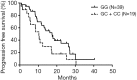
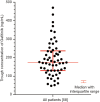
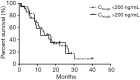
Results: Fifty-eight NSCLC patients were enrolled in this study. The median of Ctrough was 175ng/mL (range from 47.8 to 470 ng/mL). The trough concentration was not associated with either objective response or progression free survival (PFS). Ctrough was significantly lower in CYP3A4 rs2242480 CC + CT genotype than in TT genotype (P=0.019) and in ABCG2 rs2231142 AA genotype than in AC + CC genotype (P=0.031). ABCB1 rs2032582 dominant model was significantly correlated with overall response rate (ORR) and patients with GG phenotype respond better than patients with GT + TT phenotypes (84.6% vs. 51.2%, P=0.032). ABCB1 rs10256836 recessive model was significantly correlated with PFS and patients with GG phenotype achieved longer PFS than patients with GC + CC phenotypes (17.40 vs. 10.33 months, P=0.040).
Conclusions: The Ctrough of gefitinib was significantly different between CYP3A4 and ABCG2 genotypes, but not with the efficacy of gefitinib treatment. ABCB1 rs2032582 and rs10256836 polymorphisms were correlated treatment outcome. Polymorphisms analysis of ABCB1 could be a predictive biomarker for gefitinib treatment.
Keywords: ABCB1; Non-small cell lung cancer (NSCLC); gefitinib; pharmacokinetic; single nucleotide polymorphisms (SNPs).
2019 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figures



Similar articles
-
The impact of ABCB1, CYP3A4/5 and ABCG2 gene polymorphisms on rivaroxaban trough concentrations and bleeding events in patients with non-valvular atrial fibrillation.Hum Genomics. 2023 Jul 7;17(1):59. doi: 10.1186/s40246-023-00506-3. Hum Genomics. 2023. PMID: 37420302 Free PMC article.
-
Effects of p450 Polymorphisms on the Clinical Outcomes of Gefitinib Treatment in Patients with Epidermal Growth Factor Receptor Mutation-Positive Non-Small Cell Lung Cancer.Genet Test Mol Biomarkers. 2022 Dec;26(12):582-588. doi: 10.1089/gtmb.2022.0070. Genet Test Mol Biomarkers. 2022. PMID: 36577124
-
Association of pharmacokinetics and pharmacogenomics with safety and efficacy of gefitinib in patients with EGFR mutation positive advanced non-small cell lung cancer.Lung Cancer. 2016 Mar;93:69-76. doi: 10.1016/j.lungcan.2016.01.005. Epub 2016 Jan 11. Lung Cancer. 2016. PMID: 26898617
-
Role of EGFR SNPs in survival of advanced lung adenocarcinoma patients treated with Gefitinib.Gene. 2013 Mar 15;517(1):60-4. doi: 10.1016/j.gene.2012.12.087. Epub 2013 Jan 9. Gene. 2013. PMID: 23313300
-
Effect of CYP3A and ABCB1 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of calcineurin inhibitors: Part I.Clin Pharmacokinet. 2010 Mar;49(3):141-75. doi: 10.2165/11317350-000000000-00000. Clin Pharmacokinet. 2010. PMID: 20170205 Review.
Cited by
-
Establishment and application of a predictive model for gefitinib-induced severe rash based on pharmacometabolomic profiling and polymorphisms of transporters in non-small cell lung cancer.Transl Oncol. 2021 Jan;14(1):100951. doi: 10.1016/j.tranon.2020.100951. Epub 2020 Nov 19. Transl Oncol. 2021. PMID: 33221684 Free PMC article.
-
The role of salvage surgery in the treatment of a gefitinib-resistant non-small cell lung cancer patient: a case report.J Thorac Dis. 2021 Jul;13(7):4554-4559. doi: 10.21037/jtd-21-171. J Thorac Dis. 2021. PMID: 34422381 Free PMC article. No abstract available.
-
Pharmacokinetics, safety, tolerability, and feasibility of apatinib in combination with gefitinib in stage IIIB-IV EGFR-mutated non-squamous NSCLC: a drug-drug interaction study.Cancer Chemother Pharmacol. 2023 Nov;92(5):411-418. doi: 10.1007/s00280-023-04563-2. Epub 2023 Jul 31. Cancer Chemother Pharmacol. 2023. PMID: 37518060
-
Lipidomics reveals that sustained SREBP-1-dependent lipogenesis is a key mediator of gefitinib-acquired resistance in EGFR-mutant lung cancer.Cell Death Discov. 2021 Nov 13;7(1):353. doi: 10.1038/s41420-021-00744-1. Cell Death Discov. 2021. PMID: 34775471 Free PMC article.
-
Different efficacy in the non-small cell lung cancer patient with bilateral synchronous lesions treated with neoadjuvant gefitinib therapy: a case report.J Thorac Dis. 2020 Apr;12(4):1582-1587. doi: 10.21037/jtd.2020.02.60. J Thorac Dis. 2020. PMID: 32395295 Free PMC article.
References
-
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. 10.1016/S1470-2045(09)70364-X - DOI - PubMed
-
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. 10.1016/S2213-2600(18)30264-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
