In silico analysis for determining the deleterious nonsynonymous single nucleotide polymorphisms of BRCA genes
- PMID: 32042831
- PMCID: PMC6995337
- DOI: 10.22099/mbrc.2019.34198.1420
In silico analysis for determining the deleterious nonsynonymous single nucleotide polymorphisms of BRCA genes
Abstract
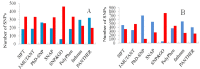
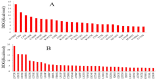
Recent advances in DNA sequencing techniques have led to an increase in the identification of single nucleotide polymorphisms (SNPs) in BRCA1 and BRCA2 genes, but no further information regarding the deleterious probability of many of them is available (Variants of Unknown Significance/VUS). As a result, in the current study, different sequence- and structure-based computational tools including SIFT, PolyPhen2, PANTHER, SNPs&GO, FATHMM, SNAP, PhD-SNP, Align-GVGD, and I-Mutant were utilized for determining how resulted BRCA protein is affected by corresponding missense mutations. FoldX was used to estimate mutational effects on the structural stability of BRCA proteins. Variants were considered extremely deleterious only when all tools predicted them to be deleterious. A total of 10 VUSs in BRCA1 (Cys39Ser, Cys64Gly, Phe861Cys, Arg1699Pro, Trp1718Cys, Phe1761Ser, Gly1788Asp, Val1804Gly, Trp1837Gly, and Trp1837Cys) and 12 in BRCA2 (Leu2510Pro, Asp2611Gly, Tyr2660Asp, Leu2686Pro, Leu2688Pro, Tyr2726Cys, Leu2792Pro, Gly2812Glu, Gly2813Glu, Arg2842Cys, Asp3073Gly, and Gly3076Val) were considered as extremely deleterious. Results suggested that deleterious variants were mostly enriched in the N- and C-terminal domain of the BRCA1 and BRCA2 C-terminus. Utilizing evolutionary conservation analysis, we demonstrated that the majority of deleterious SNPs ensue in highly conserved regions of BRCA genes. Furthermore, utilizing FoldX, we demonstrated that alterations in the function of proteins are not always together with stability alterations.
Keywords: BRCA genes; SNPs; computational tools; evolutionary analysis.
Conflict of interest statement
Authors have no competing interests
Figures

References
-
- Hopper JL, Southey MC, Dite GS, Jolley DJ, Giles GG, McCredie MR, Easton DF, Venter DJ. Population-based estimate of the average age-specific cumulative risk of breast cancer for a defined set of protein-truncating mutations in BRCA1 and BRCA2. Cancer Epidem Biomar. 1999;8:741–747. - PubMed
-
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. - PubMed
-
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. - PubMed
-
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products. .. Functional motifs... Nat Genet. 1996;13:266–268. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
