An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida
- PMID: 32045111
- PMCID: PMC7017828
- DOI: 10.1111/1751-7915.13533
An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida
Abstract
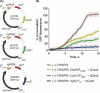
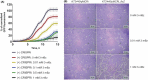
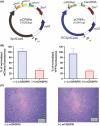
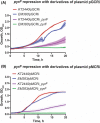
Owing to its wide metabolic versatility and physiological robustness, together with amenability to genetic manipulations and high resistance to stressful conditions, Pseudomonas putida is increasingly becoming the organism of choice for a range of applications in both industrial and environmental applications. However, a range of applied synthetic biology and metabolic engineering approaches are still limited by the lack of specific genetic tools to effectively and efficiently regulate the expression of target genes. Here, we present a single-plasmid CRISPR-interference (CRISPRi) system expressing a nuclease-deficient cas9 gene under the control of the inducible XylS/Pm expression system, along with the option of adopting constitutively expressed guide RNAs (either sgRNA or crRNA and tracrRNA). We showed that the system enables tunable, tightly controlled gene repression (up to 90%) of chromosomally expressed genes encoding fluorescent proteins, either individually or simultaneously. In addition, we demonstrate that this method allows for suppressing the expression of the essential genes pyrF and ftsZ, resulting in significantly low growth rates or morphological changes respectively. This versatile system expands the capabilities of the current CRISPRi toolbox for efficient, targeted and controllable manipulation of gene expression in P. putida.
© 2020 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Akkaya, Ö. , Nikel, P.I. , Pérez‐Pantoja, D. , and de Lorenzo, V. (2019) Evolving metabolism of 2,4‐dinitrotoluene triggers SOS‐independent diversification of host cells. Environ Microbiol 21: 314–326. - PubMed
-
- Aparicio, T. , de Lorenzo, V. , and Martínez‐García, E. (2018) CRISPR/Cas9‐based counterselection boosts recombineering efficiency in Pseudomonas putida . Biotechnol J 13: 1700161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

