A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model
- PMID: 32046307
- PMCID: PMC7157757
- DOI: 10.3390/vaccines8010077
A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model
Expression of concern in
-
Expression of Concern: Artigas-Jerónimo et al. A Novel Combined Scientific and Artistic Approach for the Advanced Characterization of Interactomes: The Akirin/Subolesin Model. Vaccines 2020, 8, 77.Vaccines (Basel). 2020 Jul 25;8(3):418. doi: 10.3390/vaccines8030418. Vaccines (Basel). 2020. PMID: 32722443 Free PMC article.
Abstract
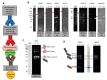
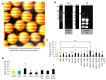
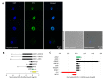
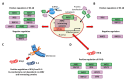
The main objective of this study was to propose a novel methodology to approach challenges in molecular biology. Akirin/Subolesin (AKR/SUB) are vaccine protective antigens and are a model for the study of the interactome due to its conserved function in the regulation of different biological processes such as immunity and development throughout the metazoan. Herein, three visual artists and a music professor collaborated with scientists for the functional characterization of the AKR2 interactome in the regulation of the NF-κB pathway in human placenta cells. The results served as a methodological proof-of-concept to advance this research area. The results showed new perspectives on unexplored characteristics of AKR2 with functional implications. These results included protein dimerization, the physical interactions with different proteins simultaneously to regulate various biological processes defined by cell type-specific AKR-protein interactions, and how these interactions positively or negatively regulate the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway in a biological context-dependent manner. These results suggested that AKR2-interacting proteins might constitute suitable secondary transcription factors for cell- and stimulus-specific regulation of NF-κB. Musical perspective supported AKR/SUB evolutionary conservation in different species and provided new mechanistic insights into the AKR2 interactome. The combined scientific and artistic perspectives resulted in a multidisciplinary approach, advancing our knowledge on AKR/SUB interactome, and provided new insights into the function of AKR2-protein interactions in the regulation of the NF-κB pathway. Additionally, herein we proposed an algorithm for quantum vaccinomics by focusing on the model proteins AKR/SUB.
Keywords: NF-κB; akirin; art; evolution; interactome; music; protective epitope; quantum vaccinomics; subolesin; vaccine; yeast two-hybrid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Jeffries S. When two tribes meet: Collaborations between artists and scientists. [(accessed on 1 August 2019)];The Guardian. 2011 Aug 21; Available online: https://www.theguardian.com/artanddesign/2011/aug/21/collaborations-betw....
-
- Tayag Y., Wells B. Art and evolution: A work in progress. [(accessed on 1 August 2019)];Scientific American. 2014 May 6; Available online: https://blogs.scientificamerican.com/guest-blog/art-and-evolution-a-work...
-
- Veis N. Four takes in the evolution of art. Nature. 2017;543:490. doi: 10.1038/543490a. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

