Trait differentiation and modular toxin expression in palm-pitvipers
- PMID: 32046632
- PMCID: PMC7014597
- DOI: 10.1186/s12864-020-6545-9
Trait differentiation and modular toxin expression in palm-pitvipers
Abstract
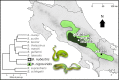
Background: Modularity is the tendency for systems to organize into semi-independent units and can be a key to the evolution and diversification of complex biological systems. Snake venoms are highly variable modular systems that exhibit extreme diversification even across very short time scales. One well-studied venom phenotype dichotomy is a trade-off between neurotoxicity versus hemotoxicity that occurs through the high expression of a heterodimeric neurotoxic phospholipase A2 (PLA2) or snake venom metalloproteinases (SVMPs). We tested whether the variation in these venom phenotypes could occur via variation in regulatory sub-modules through comparative venom gland transcriptomics of representative Black-Speckled Palm-Pitvipers (Bothriechis nigroviridis) and Talamancan Palm-Pitvipers (B. nubestris).
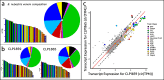
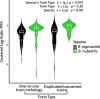
Results: We assembled 1517 coding sequences, including 43 toxins for B. nigroviridis and 1787 coding sequences including 42 toxins for B. nubestris. The venom gland transcriptomes were extremely divergent between these two species with one B. nigroviridis exhibiting a primarily neurotoxic pattern of expression, both B. nubestris expressing primarily hemorrhagic toxins, and a second B. nigroviridis exhibiting a mixed expression phenotype. Weighted gene coexpression analyses identified six submodules of transcript expression variation, one of which was highly associated with SVMPs and a second which contained both subunits of the neurotoxic PLA2 complex. The sub-module association of these toxins suggest common regulatory pathways underlie the variation in their expression and is consistent with known patterns of inheritance of similar haplotypes in other species. We also find evidence that module associated toxin families show fewer gene duplications and transcript losses between species, but module association did not appear to affect sequence diversification.
Conclusion: Sub-modular regulation of expression likely contributes to the diversification of venom phenotypes within and among species and underscores the role of modularity in facilitating rapid evolution of complex traits.
Keywords: Bothriechis; Gene family evolution; Modularity; Transcriptomics; Venom.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

