Comparative genome characterization of the periodontal pathogen Tannerella forsythia
- PMID: 32046654
- PMCID: PMC7014623
- DOI: 10.1186/s12864-020-6535-y
Comparative genome characterization of the periodontal pathogen Tannerella forsythia
Abstract
Background: Tannerella forsythia is a bacterial pathogen implicated in periodontal disease. Numerous virulence-associated T. forsythia genes have been described, however, it is necessary to expand the knowledge on T. forsythia's genome structure and genetic repertoire to further elucidate its role within pathogenesis. Tannerella sp. BU063, a putative periodontal health-associated sister taxon and closest known relative to T. forsythia is available for comparative analyses. In the past, strain confusion involving the T. forsythia reference type strain ATCC 43037 led to discrepancies between results obtained from in silico analyses and wet-lab experimentation.
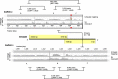
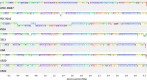
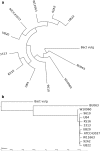
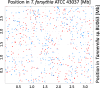
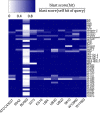
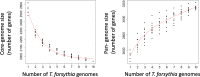
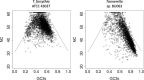
Results: We generated a substantially improved genome assembly of T. forsythia ATCC 43037 covering 99% of the genome in three sequences. Using annotated genomes of ten Tannerella strains we established a soft core genome encompassing 2108 genes, based on orthologs present in > = 80% of the strains analysed. We used a set of known and hypothetical virulence factors for comparisons in pathogenic strains and the putative periodontal health-associated isolate Tannerella sp. BU063 to identify candidate genes promoting T. forsythia's pathogenesis. Searching for pathogenicity islands we detected 38 candidate regions in the T. forsythia genome. Only four of these regions corresponded to previously described pathogenicity islands. While the general protein O-glycosylation gene cluster of T. forsythia ATCC 43037 has been described previously, genes required for the initiation of glycan synthesis are yet to be discovered. We found six putative glycosylation loci which were only partially conserved in other bacteria. Lastly, we performed a comparative analysis of translational bias in T. forsythia and Tannerella sp. BU063 and detected highly biased genes.
Conclusions: We provide resources and important information on the genomes of Tannerella strains. Comparative analyses enabled us to assess the suitability of T. forsythia virulence factors as therapeutic targets and to suggest novel putative virulence factors. Further, we report on gene loci that should be addressed in the context of elucidating T. forsythia's protein O-glycosylation pathway. In summary, our work paves the way for further molecular dissection of T. forsythia biology in general and virulence of this species in particular.
Keywords: Codon usage bias; Comparative genomics; Computational analysis; Genome assembly; Glycosylation gene cluster; Pan-genome; Pathogenicity island; Periodontitis; Tannerella; Virulence.
Conflict of interest statement
HH is a member of the editorial board of BMC Genomics.
Figures
Similar articles
-
A General Protein O-Glycosylation Gene Cluster Encodes the Species-Specific Glycan of the Oral Pathogen Tannerella forsythia: O-Glycan Biosynthesis and Immunological Implications.Front Microbiol. 2018 Aug 28;9:2008. doi: 10.3389/fmicb.2018.02008. eCollection 2018. Front Microbiol. 2018. PMID: 30210478 Free PMC article.
-
Single cell genomics of uncultured, health-associated Tannerella BU063 (Oral Taxon 286) and comparison to the closely related pathogen Tannerella forsythia.PLoS One. 2014 Feb 14;9(2):e89398. doi: 10.1371/journal.pone.0089398. eCollection 2014. PLoS One. 2014. PMID: 24551246 Free PMC article.
-
Characterization of the O-Glycoproteome of Tannerella forsythia.mSphere. 2021 Oct 27;6(5):e0064921. doi: 10.1128/mSphere.00649-21. Epub 2021 Sep 15. mSphere. 2021. PMID: 34523981 Free PMC article.
-
Nonulosonic acids contribute to the pathogenicity of the oral bacterium Tannerella forsythia.Interface Focus. 2019 Apr 6;9(2):20180064. doi: 10.1098/rsfs.2018.0064. Epub 2019 Feb 15. Interface Focus. 2019. PMID: 30842870 Free PMC article. Review.
-
Peptidoglycan Salvage Enables the Periodontal Pathogen Tannerella forsythia to Survive within the Oral Microbial Community.Microb Physiol. 2021;31(2):123-134. doi: 10.1159/000516751. Epub 2021 Jun 9. Microb Physiol. 2021. PMID: 34107471 Review.
Cited by
-
Genomic Analysis of 18th-Century Kazakh Individuals and Their Oral Microbiome.Biology (Basel). 2021 Dec 14;10(12):1324. doi: 10.3390/biology10121324. Biology (Basel). 2021. PMID: 34943238 Free PMC article.
-
Metabolomics Research in Periodontal Disease by Mass Spectrometry.Molecules. 2022 Apr 30;27(9):2864. doi: 10.3390/molecules27092864. Molecules. 2022. PMID: 35566216 Free PMC article. Review.
-
Analysis of oral microbiome from fossil human remains revealed the significant differences in virulence factors of modern and ancient Tannerella forsythia.BMC Genomics. 2020 Jun 15;21(1):402. doi: 10.1186/s12864-020-06810-9. BMC Genomics. 2020. PMID: 32539695 Free PMC article.
-
Changes in the oral status and periodontal pathogens in a Sardinian rural community from pre-industrial to modern time.Sci Rep. 2022 Sep 23;12(1):15895. doi: 10.1038/s41598-022-20193-9. Sci Rep. 2022. PMID: 36151274 Free PMC article.
-
Myocardial infarction risk is increased by periodontal pathobionts: a cross-sectional study.Sci Rep. 2022 Nov 3;12(1):18608. doi: 10.1038/s41598-022-19154-z. Sci Rep. 2022. PMID: 36329042 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

