Effect of the TetR family transcriptional regulator Sp1418 on the global metabolic network of Saccharopolyspora pogona
- PMID: 32046731
- PMCID: PMC7011500
- DOI: 10.1186/s12934-020-01299-z
Effect of the TetR family transcriptional regulator Sp1418 on the global metabolic network of Saccharopolyspora pogona
Abstract
Background: Saccharopolyspora pogona is a prominent industrial strain due to its production of butenyl-spinosyn, a high-quality insecticide against a broad spectrum of insect pests. TetR family proteins are diverse in a tremendous number of microorganisms and some are been researched to have a key role in metabolic regulation. However, specific functions of TetR family proteins in S. pogona are yet to characterize.
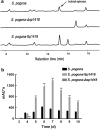
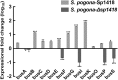
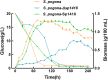
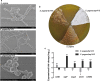
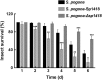
Results: In the present study, the overexpression of the tetR-like gene sp1418 in S. pogona resulted in marked effects on vegetative growth, sporulation, butenyl-spinosyn biosynthesis, and oxidative stress. By using qRT-PCR analysis, mass spectrometry, enzyme activity detection, and sp1418 knockout verification, we showed that most of these effects could be attributed to the overexpression of Sp1418, which modulated enzymes related to the primary metabolism, oxidative stress and secondary metabolism, and thereby resulted in distinct growth characteristics and an unbalanced supply of precursor monomers for butenyl-spinosyn biosynthesis.
Conclusion: This study revealed the function of Sp1418 and enhanced the understanding of the metabolic network in S. pogona, and provided insights into the improvement of secondary metabolite production.
Keywords: Butenyl-spinosyn; Oxidative stress; Saccharopolyspora pogona; TetR family transcriptional regulator.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Microb Cell Fact. 2021 Aug 14;20(1):157. doi: 10.1186/s12934-021-01651-x. Microb Cell Fact. 2021. PMID: 34391414 Free PMC article.
-
Identification of a TetR family regulator and a polyketide synthase gene cluster involved in growth development and butenyl-spinosyn biosynthesis of Saccharopolyspora pogona.Appl Microbiol Biotechnol. 2021 Feb;105(4):1519-1533. doi: 10.1007/s00253-021-11105-4. Epub 2021 Jan 23. Appl Microbiol Biotechnol. 2021. PMID: 33484320
-
A TetR family transcriptional regulator, SP_2854 can affect the butenyl-spinosyn biosynthesis by regulating glucose metabolism in Saccharopolyspora pogona.Microb Cell Fact. 2022 May 14;21(1):83. doi: 10.1186/s12934-022-01808-2. Microb Cell Fact. 2022. PMID: 35568948 Free PMC article.
-
Recent advances in the biochemistry of spinosyns.Appl Microbiol Biotechnol. 2009 Feb;82(1):13-23. doi: 10.1007/s00253-008-1784-8. Epub 2008 Dec 10. Appl Microbiol Biotechnol. 2009. PMID: 19082588 Review.
-
Genes for the biosynthesis of spinosyns: applications for yield improvement in Saccharopolyspora spinosa.J Ind Microbiol Biotechnol. 2001 Dec;27(6):399-402. doi: 10.1038/sj.jim.7000180. J Ind Microbiol Biotechnol. 2001. PMID: 11774006 Review.
Cited by
-
Effects of a Pirin-like protein on strain growth and spinosad biosynthesis in Saccharopolyspora spinosa.Appl Microbiol Biotechnol. 2023 Sep;107(17):5439-5451. doi: 10.1007/s00253-023-12636-8. Epub 2023 Jul 10. Appl Microbiol Biotechnol. 2023. PMID: 37428187
-
Inactivation of Nitrite-Dependent Nitric Oxide Biosynthesis Is Responsible for Overlapped Antibiotic Resistance between Naturally and Artificially Evolved Pseudomonas aeruginosa.mSystems. 2021 Oct 26;6(5):e0073221. doi: 10.1128/mSystems.00732-21. Epub 2021 Sep 21. mSystems. 2021. PMID: 34546070 Free PMC article.
-
Improving butenyl-spinosyn production in Saccharopolyspora pogona through combination of metabolic engineering and medium optimization.Front Microbiol. 2025 Apr 23;16:1561042. doi: 10.3389/fmicb.2025.1561042. eCollection 2025. Front Microbiol. 2025. PMID: 40336833 Free PMC article.
-
Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Microb Cell Fact. 2021 Aug 14;20(1):157. doi: 10.1186/s12934-021-01651-x. Microb Cell Fact. 2021. PMID: 34391414 Free PMC article.
-
The PurR family transcriptional regulator promotes butenyl-spinosyn production in Saccharopolyspora pogona.Appl Microbiol Biotechnol. 2025 Jan 21;109(1):14. doi: 10.1007/s00253-024-13390-1. Appl Microbiol Biotechnol. 2025. PMID: 39836216 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

