Nucleolar localization of RAG1 modulates V(D)J recombination activity
- PMID: 32047031
- PMCID: PMC7049140
- DOI: 10.1073/pnas.1920021117
Nucleolar localization of RAG1 modulates V(D)J recombination activity
Abstract
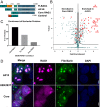
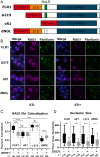
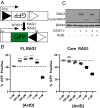
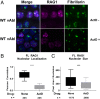
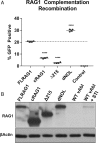
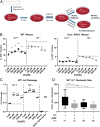
V(D)J recombination assembles and diversifies Ig and T cell receptor genes in developing B and T lymphocytes. The reaction is initiated by the RAG1-RAG2 protein complex which binds and cleaves at discrete gene segments in the antigen receptor loci. To identify mechanisms that regulate V(D)J recombination, we used proximity-dependent biotin identification to analyze the interactomes of full-length and truncated forms of RAG1 in pre-B cells. This revealed an association of RAG1 with numerous nucleolar proteins in a manner dependent on amino acids 216 to 383 and allowed identification of a motif required for nucleolar localization. Experiments in transformed pre-B cell lines and cultured primary pre-B cells reveal a strong correlation between disruption of nucleoli, reduced association of RAG1 with a nucleolar marker, and increased V(D)J recombination activity. Mutation of the RAG1 nucleolar localization motif boosts recombination while removal of the first 215 amino acids of RAG1, required for efficient egress from nucleoli, reduces recombination activity. Our findings indicate that nucleolar sequestration of RAG1 is a negative regulatory mechanism in V(D)J recombination and identify regions of the RAG1 N-terminal region that control nucleolar association and egress.
Keywords: B cell development; RAG1; V(D)J recombination; nucleolus; proximity-dependent biotin identification.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Tonegawa S., Somatic generation of antibody diversity. Nature 302, 575–581 (1983). - PubMed
-
- Schatz D. G., Oettinger M. A., Baltimore D., The V(D)J recombination activating gene, RAG-1. Cell 59, 1035–1048 (1989). - PubMed
-
- Oettinger M. A., Schatz D. G., Gorka C., Baltimore D., RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248, 1517–1523 (1990). - PubMed
-
- Schatz D. G., Ji Y., Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263 (2011). - PubMed
-
- Gellert M., V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101–132 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

