IGLV3-21 * 01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling
- PMID: 32047037
- PMCID: PMC7049113
- DOI: 10.1073/pnas.1913810117
IGLV3-21 * 01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling
Abstract
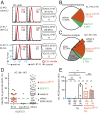
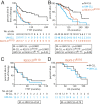
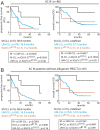
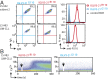
The prognosis of chronic lymphocytic leukemia (CLL) depends on different markers, including cytogenetic aberrations, oncogenic mutations, and mutational status of the immunoglobulin (Ig) heavy-chain variable (IGHV) gene. The number of IGHV mutations distinguishes mutated (M) CLL with a markedly superior prognosis from unmutated (UM) CLL cases. In addition, B cell antigen receptor (BCR) stereotypes as defined by IGHV usage and complementarity-determining regions (CDRs) classify ∼30% of CLL cases into prognostically important subsets. Subset 2 expresses a BCR with the combination of IGHV3-21-derived heavy chains (HCs) with IGLV3-21-derived light chains (LCs), and is associated with an unfavorable prognosis. Importantly, the subset 2 LC carries a single-point mutation, termed R110, at the junction between the variable and constant LC regions. By analyzing 4 independent clinical cohorts through BCR sequencing and by immunophenotyping with antibodies specifically recognizing wild-type IGLV3-21 and R110-mutated IGLV3-21 (IGLV3-21R110), we show that IGLV3-21R110-expressing CLL represents a distinct subset with poor prognosis independent of IGHV mutations. Compared with other alleles, only IGLV3-21*01 facilitates effective homotypic BCR-BCR interaction that results in autonomous, oncogenic BCR signaling after acquiring R110 as a single-point mutation. Presumably, this mutation acts as a standalone driver that transforms IGLV3-21*01-expressing B cells to develop CLL. Thus, we propose to expand the conventional definition of CLL subset 2 to subset 2L by including all IGLV3-21R110-expressing CLL cases regardless of IGHV mutational status. Moreover, the generation of monoclonal antibodies recognizing IGLV3-21 or mutated IGLV3-21R110 facilitates the recognition of B cells carrying this mutation in CLL patients or healthy donors.
Keywords: B cell antigen receptor (BCR); autonomous BCR signaling; chronic lymphocytic leukemia (CLL); immunoglobulin allele IGLV3-21*01.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: H.J. is a cofounder of AVA LifeScience GmbH that has filed patents on antibodies recognizing structures involved in autonomous BCR signaling.
Figures



References
-
- Hallek M., Chronic lymphocytic leukemia: 2015 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 90, 446–460 (2015). - PubMed
-
- Brenner H., Gondos A., Pulte D., Trends in long-term survival of patients with chronic lymphocytic leukemia from the 1980s to the early 21st century. Blood 111, 4916–4921 (2008). - PubMed
-
- Damle R. N., et al. , Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847 (1999). - PubMed
-
- Hamblin T. J., Davis Z., Gardiner A., Oscier D. G., Stevenson F. K., Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

