Structural Insights into the Plant Immune Receptors PRRs and NLRs
- PMID: 32047048
- PMCID: PMC7140948
- DOI: 10.1104/pp.19.01252
Structural Insights into the Plant Immune Receptors PRRs and NLRs
Abstract
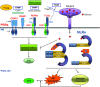
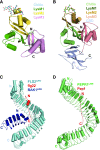
Recent progresses made in structural analysis of plant PRRs and NLRs show the advancements in cryo-EM structural biology.
Figures







References
-
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T(2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20: 47–54 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

