A Gene Cluster That Encodes Histone Deacetylase Inhibitors Contributes to Bacterial Persistence and Antibiotic Tolerance in Burkholderia thailandensis
- PMID: 32047060
- PMCID: PMC7018527
- DOI: 10.1128/mSystems.00609-19
A Gene Cluster That Encodes Histone Deacetylase Inhibitors Contributes to Bacterial Persistence and Antibiotic Tolerance in Burkholderia thailandensis
Abstract
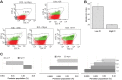
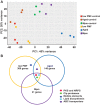
Persister cells are genetically identical variants in a bacterial population that have phenotypically modified their physiology to survive environmental stress. In bacterial pathogens, persisters are able to survive antibiotic treatment and reinfect patients in a frustrating cycle of chronic infection. To better define core persistence mechanisms for therapeutics development, we performed transcriptomics analyses of Burkholderia thailandensis populations enriched for persisters via three methods: flow sorting for low proton motive force, meropenem treatment, and culture aging. Although the three persister-enriched populations generally displayed divergent gene expression profiles that reflect the multimechanistic nature of stress adaptations, there were several common gene pathways activated in two or all three populations. These include polyketide and nonribosomal peptide synthesis, Clp proteases, mobile elements, enzymes involved in lipid metabolism, and ATP-binding cassette (ABC) transporter systems. In particular, identification of genes that encode polyketide synthases (PKSs) and fatty acid catabolism factors indicates that generation of secondary metabolites, natural products, and complex lipids could be part of the metabolic program that governs the persistence state. We also found that loss-of-function mutations in the PKS-encoding gene locus BTH_I2366, which plays a role in biosynthesis of histone deacetylase (HDAC) inhibitors, resulted in increased sensitivity to antibiotics targeting DNA replication. Furthermore, treatment of multiple bacterial pathogens with a fatty acid synthesis inhibitor, CP-640186, potentiated the efficacy of meropenem against the persister populations. Altogether, our results suggest that bacterial persisters may exhibit an outwardly dormant physiology but maintain active metabolic processes that are required to maintain persistence.IMPORTANCE The discovery of antibiotics such as penicillin and streptomycin marked a historic milestone in the 1940s and heralded a new era of antimicrobial therapy as the modern standard for medical treatment. Yet, even in those early days of discovery, it was noted that a small subset of cells (∼1 in 105) survived antibiotic treatment and continued to persist, leading to recurrence of chronic infection. These persisters are phenotypic variants that have modified their physiology to survive environmental stress. In this study, we have performed three transcriptomic screens to identify persistence genes that are common between three different stressor conditions. In particular, we identified genes that function in the synthesis of secondary metabolites, small molecules, and complex lipids, which are likely required to maintain the persistence state. Targeting universal persistence genes can lead to the development of clinically relevant antipersistence therapeutics for infectious disease management.
Keywords: Burkholderia; antibiotic resistance; bacterial persistence; stress; transcriptomics.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
