Universal Dermal Microbiome in Human Skin
- PMID: 32047129
- PMCID: PMC7018652
- DOI: 10.1128/mBio.02945-19
Universal Dermal Microbiome in Human Skin
Abstract
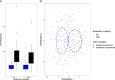
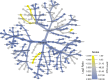
Human skin microbiota has been described as a "microbial fingerprint" due to observed differences between individuals. Current understanding of the cutaneous microbiota is based on sampling the outermost layers of the epidermis, while the microbiota in the remaining skin layers has not yet been fully characterized. Environmental conditions can vary drastically between the cutaneous compartments and give rise to unique communities. We demonstrate that the dermal microbiota is surprisingly similar among individuals and contains a specific subset of the epidermal microbiota. Variability in bacterial community composition decreased significantly from the epidermal to the dermal compartment but was similar among anatomic locations (hip and knee). The composition of the epidermal microbiota was more strongly affected by environmental factors than that of the dermal community. These results indicate a well-conserved dermal community that is functionally distinct from the epidermal community, challenging the current dogma. Future studies in cutaneous disorders and chronic infections may benefit by focusing on the dermal microbiota as a persistent microbial community.IMPORTANCE Human skin microbiota is thought to be unique according to the individual's lifestyle and genetic predisposition. This is true for the epidermal microbiota, while our findings demonstrate that the dermal microbiota is universal between healthy individuals. The preserved dermal microbial community is compositionally unique and functionally distinct to the specific environment in the depth of human skin. It is expected to have direct contact with the immune response of the human host, and research in the communication between host and microbiota should be targeted to this cutaneous compartment. This novel insight into specific microbial adaptation can be used advantageously in the research of chronic disorders and infections of the skin. It can enlighten the alteration between health and disease to the benefit of patients suffering from long-lasting socioeconomic illnesses.
Keywords: 16S rRNA genes; DNA sequencing; cutaneous compartments; dermal microbiota; dry habitat; skin biopsies; skin microbiome.
Copyright © 2020 Bay et al.
Figures

References
-
- Grice EA, NISC Comparative Sequencing Program, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographic and temporal diversity of the human skin Microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

