Both ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus nsP3 Macrodomain Affect Neurovirulence in Mice
- PMID: 32047134
- PMCID: PMC7018654
- DOI: 10.1128/mBio.03253-19
Both ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus nsP3 Macrodomain Affect Neurovirulence in Mice
Abstract
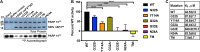
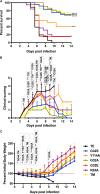
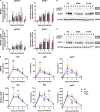
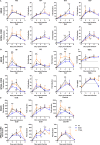
Macrodomain (MD), a highly conserved protein fold present in a subset of plus-strand RNA viruses, binds to and hydrolyzes ADP-ribose (ADPr) from ADP-ribosylated proteins. ADPr-binding by the alphavirus nonstructural protein 3 (nsP3) MD is necessary for the initiation of virus replication in neural cells, whereas hydrolase activity facilitates replication complex amplification. To determine the importance of these activities for pathogenesis of alphavirus encephalomyelitis, mutations were introduced into the nsP3 MD of Sindbis virus (SINV), and the effects on ADPr binding and hydrolase activities, virus replication, immune responses, and disease were assessed. Elimination of ADPr-binding and hydrolase activities (G32E) severely impaired in vitro replication of SINV in neural cells and in vivo replication in the central nervous systems of 2-week-old mice with reversion to wild type (WT) (G) or selection of a less compromising change (S) during replication. SINVs with decreased binding and hydrolase activities (G32S and G32A) or with hydrolase deficiency combined with better ADPr-binding (Y114A) were less virulent than WT virus. Compared to the WT, the G32S virus replicated less well in both the brain and spinal cord, induced similar innate responses, and caused less severe disease with full recovery of survivors, whereas the Y114A virus replicated well, induced higher expression of interferon-stimulated and NF-κB-induced genes, and was cleared more slowly from the spinal cord with persistent paralysis in survivors. Therefore, MD function was important for neural cell replication both in vitro and in vivo and determined the outcome from alphavirus encephalomyelitis in mice.IMPORTANCE Viral encephalomyelitis is an important cause of long-term disability, as well as acute fatal disease. Identifying viral determinants of outcome helps in assessing disease severity and developing new treatments. Mosquito-borne alphaviruses infect neurons and cause fatal disease in mice. The highly conserved macrodomain of nonstructural protein 3 binds and can remove ADP-ribose (ADPr) from ADP-ribosylated proteins. To determine the importance of these functions for virulence, recombinant mutant viruses were produced. If macrodomain mutations eliminated ADPr-binding or hydrolase activity, viruses did not grow. If the binding and hydrolase activities were impaired, the viruses grew less well than the wild-type virus, induced similar innate responses, and caused less severe disease, and most of the infected mice recovered. If binding was improved, but hydrolase activity was decreased, the virus replicated well and induced greater innate responses than did the WT, but clearance from the nervous system was impaired, and mice remained paralyzed. Therefore, macrodomain function determined the outcome of alphavirus encephalomyelitis.
Keywords: ADP-ribosyl hydrolase; ADP-ribosyl-binding activity; Sindbis virus; alphavirus; antibody; encephalomyelitis; innate immune response; macrodomain; nsP3.
Copyright © 2020 Abraham et al.
Figures


References
-
- Sane J, Kurkela S, Desdouits M, Kalimo H, Mazalrey S, Lokki ML, Vaheri A, Helve T, Tornwall J, Huerre M, Butler-Browne G, Ceccaldi PE, Gessain A, Vapalahti O. 2012. Prolonged myalgia in Sindbis virus infection: case description and in vitro infection of myotubes and myoblasts. J Infect Dis 206:407–414. doi:10.1093/infdis/jis358. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
