Genome Dynamics of Vibrio cholerae Isolates Linked to Seasonal Outbreaks of Cholera in Dhaka, Bangladesh
- PMID: 32047137
- PMCID: PMC7018647
- DOI: 10.1128/mBio.03339-19
Genome Dynamics of Vibrio cholerae Isolates Linked to Seasonal Outbreaks of Cholera in Dhaka, Bangladesh
Abstract
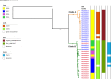
The temporal switching of serotypes from serotype Ogawa to Inaba and back to Ogawa was identified in Vibrio cholerae O1, which was responsible for seasonal outbreaks of cholera in Dhaka during the period 2015 to 2018. In order to delineate the factors responsible for this serotype transition, we performed whole-genome sequencing (WGS) of V. cholerae O1 multidrug-resistant strains belonging to both the serotypes that were isolated during this interval where the emergence and subsequent reduction of the Inaba serotype occurred. The whole-genome-based phylogenetic analysis revealed clonal expansion of the Inaba isolates mainly responsible for the peaks of infection during 2016 to 2017 and that they might have evolved from the prevailing Ogawa strains in 2015 which coclustered with them. Furthermore, the wbeT gene in these Inaba serotype isolates was inactivated due to insertion of a transposable element at the same position signifying the clonal expansion. Also, V. cholerae isolates in the Inaba serotype dominant clade mainly contained classical ctxB allele and revealed differences in the genetic composition of Vibrio
Keywords: Vibrio cholerae; genomics; seasonality; serogroups.
Copyright © 2020 Baddam et al.
Figures



References
-
- Marin MA, Thompson CC, Freitas FS, Fonseca EL, Aboderin AO, Zailani SB, Quartey NKE, Okeke IN, Vicente A. 2013. Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS Negl Trop Dis 7:e2049. doi: 10.1371/journal.pntd.0002049. - DOI - PMC - PubMed
-
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JLN, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. - DOI - PMC - PubMed

