Non-secretory renin reduces oxidative stress and increases cardiomyoblast survival during glucose and oxygen deprivation
- PMID: 32047214
- PMCID: PMC7012910
- DOI: 10.1038/s41598-020-59216-8
Non-secretory renin reduces oxidative stress and increases cardiomyoblast survival during glucose and oxygen deprivation
Abstract
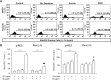
Although the renin-angiotensin system usually promotes oxidative stress and cell death, renin transcripts have been discovered, whose transcription product may be cardioprotective. These transcripts encode a non-secretory renin isoform that is localized in the cytosol and within mitochondria. Here we tested the hypotheses that cytosolic renin [ren(2-9)] expression promotes cell survival under hypoxia and glucose depletion by preserving the mitochondrial membrane potential (∆Ψm) and mitigating the accumulation of ROS. To simulate ischemic insults, we exposed H9c2 cells to glucose deprivation, anoxia or to combined oxygen-glucose deprivation (OGD) for 24 hours and determined renin expression. Furthermore, H9c2 cells transfected with the empty pIRES vector (pIRES cells) or ren(2-9) cDNA-containing vector [ren(2-9) cells] were analyzed for cell death, ∆Ψm, ATP levels, accumulation of ROS, and cytosolic Ca2+ content. In pIRES cells, expression of ren(1A-9) was stimulated under all three ischemia-related conditions. After OGD, the cells lost their ∆Ψm and exhibited enhanced ROS accumulation, increased cytosolic Ca2+ levels, decreased ATP levels as well as increased cell death. In contrast, ren(2-9) cells were markedly protected from these effects. Ren(2-9) appears to represent a protective response to OGD by reducing ROS generation and preserving mitochondrial functions. Therefore, it is a promising new target for the prevention of ischemia-induced myocardial damage.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

