Mercury 4.0: from visualization to analysis, design and prediction
- PMID: 32047413
- PMCID: PMC6998782
- DOI: 10.1107/S1600576719014092
Mercury 4.0: from visualization to analysis, design and prediction
Abstract
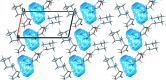
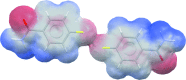
The program Mercury, developed at the Cambridge Crystallographic Data Centre, was originally designed primarily as a crystal structure visualization tool. Over the years the fields and scientific communities of chemical crystallography and crystal engineering have developed to require more advanced structural analysis software. Mercury has evolved alongside these scientific communities and is now a powerful analysis, design and prediction platform which goes a lot further than simple structure visualization.
Keywords: Mercury; computer programs; crystal packing; crystal structure visualization; structure comparison.
© Clare F. Macrae et al. 2020.
Figures






References
-
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. - PubMed
-
- Allen, F. H., Johnson, O., Shields, G. P., Smith, B. R. & Towler, M. (2004). J. Appl. Cryst. 37, 335–338.
-
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. - PubMed
-
- Bruno, I. J., Cole, J. C., Kessler, M., Luo, J., Motherwell, W. D. S., Purkis, L. H., Smith, B. R., Taylor, R., Cooper, R. I., Harris, S. E. & Orpen, A. G. (2004). J. Chem. Inf. Comput. Sci. 44, 2133–2144. - PubMed
-
- Bruno, I. J., Cole, J. C., Lommerse, J. P. M., Rowland, R. S., Taylor, R. & Verdonk, M. L. (1997). J. Comput. Aided Mol. Des. 11, 525–537. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
