Myxobacterial Response to Methyljasmonate Exposure Indicates Contribution to Plant Recruitment of Micropredators
- PMID: 32047489
- PMCID: PMC6997564
- DOI: 10.3389/fmicb.2020.00034
Myxobacterial Response to Methyljasmonate Exposure Indicates Contribution to Plant Recruitment of Micropredators
Abstract
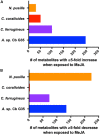
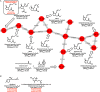
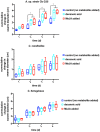
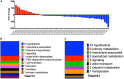
Chemical exchanges between plants and microbes within rhizobiomes are critical to the development of community structure. Volatile root exudates such as the phytohormone methyljasmonate (MeJA) contribute to various plant stress responses and have been implicated to play a role in the maintenance of microbial communities. Myxobacteria are competent predators of plant pathogens and are generally considered beneficial to rhizobiomes. While plant recruitment of myxobacteria to stave off pathogens has been suggested, no involved chemical signaling processes are known. Herein we expose predatory myxobacteria to MeJA and employ untargeted mass spectrometry, motility assays, and RNA sequencing to monitor changes in features associated with predation such as specialized metabolism, swarm expansion, and production of lytic enzymes. From a panel of four myxobacteria, we observe the most robust metabolic response from plant-associated Archangium sp. strain Cb G35 with 10 μM MeJA impacting the production of at least 300 metabolites and inducing a ≥ fourfold change in transcription for 56 genes. We also observe that MeJA induces A. sp. motility supporting plant recruitment of a subset of the investigated micropredators. Provided the varying responses to MeJA exposure, our observations indicate that MeJA contributes to the recruitment of select predatory myxobacteria suggesting further efforts are required to explore the microbial impact of plant exudates associated with biotic stress.
Keywords: methyljasmonate; micropredator; myxobacteria; phytohormones; rhizobiome.
Copyright © 2020 Adaikpoh, Akbar, Albataineh, Misra, Sharp and Stevens.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

