LRG1 Suppresses Migration and Invasion of Esophageal Squamous Cell Carcinoma by Modulating Epithelial to Mesenchymal Transition
- PMID: 32047555
- PMCID: PMC6995366
- DOI: 10.7150/jca.36189
LRG1 Suppresses Migration and Invasion of Esophageal Squamous Cell Carcinoma by Modulating Epithelial to Mesenchymal Transition
Abstract
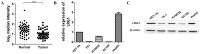
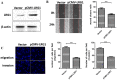
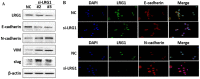
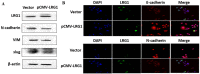
Background: Esophageal squamous cell carcinoma (ESCC) is a common cancer with poor prognosis. The molecular pathogenesis underlying ESCC remains to be explored. Leucine-rich ɑ-2-glycoprotein 1 (LRG1) has been implicated in the pathogenesis of various cancer types, however its role in ESCC is unknown. Materials and Methods: Data from the public database was analyzed to address the expression of LRG1 in ESCC. Gain-of-function studies were performed in select ESCC cell lines by over-expression or addition of recombinant LRG1, while loss-of-function studies achieved by small interfering RNA mediated knockdown. Wound healing and transwell assays were conducted to investigate ESCC cell migration and invasion upon manipulating LRG1 levels. Western blot and Immunofluorescence staining were used to examine the changes in epithelial to mesenchymal transition (EMT) and TGFβ signaling pathway. Results: LRG1 mRNA levels were found to be significantly down-regulated in patients with ESCC as well as in several ESCC cell lines. Silencing of LRG1 promoted, while overexpression of LRG1 inhibited ESCC cell migration and invasion. In line with this, Silencing of LRG1 enhanced, while overexpression of LRG1 reduced TGFβ signaling and EMT of ESCC cells. Conclusion/Significance: LRG1 suppresses ESCC cell migration and invasion via negative modulation of TGFβ signaling and EMT. Down-regulation of LRG1 in ESCC patients may favor tumor metastasis and disease progression.
Keywords: Epithelial to Mesenchymal Transition; Esophageal Squamous Cell Carcinoma; Invasion; LRG1; Migration; TGFβ signaling.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
LRG1 in cancer: mechanisms, potential therapeutic target, and use in clinical diagnosis.Mol Biol Rep. 2025 Jun 10;52(1):577. doi: 10.1007/s11033-025-10669-y. Mol Biol Rep. 2025. PMID: 40493117 Review.
-
MiR-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway.Cancer Biomark. 2017 Dec 6;20(4):527-537. doi: 10.3233/CBM-170345. Cancer Biomark. 2017. PMID: 28800315
-
The Clinical Prognostic Value of LRG1 in Esophageal Squamous Cell Carcinoma.Curr Cancer Drug Targets. 2019;19(9):756-763. doi: 10.2174/1568009619666190204095942. Curr Cancer Drug Targets. 2019. PMID: 30714525
-
Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma.J Exp Clin Cancer Res. 2017 Nov 16;36(1):161. doi: 10.1186/s13046-017-0622-1. J Exp Clin Cancer Res. 2017. PMID: 29145896 Free PMC article.
-
LRG1: an emerging player in disease pathogenesis.J Biomed Sci. 2022 Jan 21;29(1):6. doi: 10.1186/s12929-022-00790-6. J Biomed Sci. 2022. PMID: 35062948 Free PMC article. Review.
Cited by
-
Quantitative proteomics analysis of papillary thyroid carcinoma reveals protein S, clusterin, and leucine-rich α-2-glycoprotein 1 as potential prognostic protein biomarkers.Medicine (Baltimore). 2025 Aug 8;104(32):e43715. doi: 10.1097/MD.0000000000043715. Medicine (Baltimore). 2025. PMID: 40797389 Free PMC article.
-
Leucine rich alpha-2-glycoprotein 1 (Lrg1) silencing protects against sepsis-mediated brain injury by inhibiting transforming growth factor beta1 (TGFβ1)/SMAD signaling pathway.Bioengineered. 2022 Mar;13(3):7316-7327. doi: 10.1080/21655979.2022.2048775. Bioengineered. 2022. PMID: 35264055 Free PMC article.
-
SUMOylation of HSP27 regulates PKM2 to promote esophageal squamous cell carcinoma progression.Oncol Rep. 2020 Oct;44(4):1355-1364. doi: 10.3892/or.2020.7711. Epub 2020 Jul 31. Oncol Rep. 2020. PMID: 32945483 Free PMC article.
-
LRG-1 promotes fat graft survival through the RAB31-mediated inhibition of hypoxia-induced apoptosis.J Cell Mol Med. 2022 Jun;26(11):3153-3168. doi: 10.1111/jcmm.17280. Epub 2022 Mar 23. J Cell Mol Med. 2022. PMID: 35322540 Free PMC article.
-
LRG1 in cancer: mechanisms, potential therapeutic target, and use in clinical diagnosis.Mol Biol Rep. 2025 Jun 10;52(1):577. doi: 10.1007/s11033-025-10669-y. Mol Biol Rep. 2025. PMID: 40493117 Review.
References
-
- Di Pardo BJ, Bronson NW, Diggs BS, Thomas CR Jr, Hunter JG, Dolan JP. The Global Burden of Esophageal Cancer: A Disability-Adjusted Life-Year Approach. World J Surg. 2016;40:395–401. - PubMed
-
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–509. - PubMed
-
- Song Y, Li L, Ou Y, Gao Z, Li E, Li X. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

