IFN-γ establishes interferon-stimulated gene-mediated antiviral state against Newcastle disease virus in chicken fibroblasts
- PMID: 32047904
- PMCID: PMC7109688
- DOI: 10.1093/abbs/gmz158
IFN-γ establishes interferon-stimulated gene-mediated antiviral state against Newcastle disease virus in chicken fibroblasts
Abstract
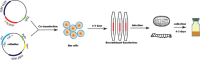
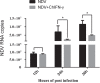
Newcastle disease virus (NDV) causes severe economic losses through severe morbidity and mortality and poses a significant threat to the global poultry industry. Significant efforts have been made to develop novel vaccines and therapeutics; however, the interaction of NDV with the host is not yet fully understood. Interferons (IFNs), an integral component of innate immune signaling, act as the first line of defense against invading viruses. Compared with the mammalian repertoire of IFNs, limited information is available on the antiviral potential of IFNs in chickens. Here, we expressed chicken IFN-γ (chIFN-γ) using a baculovirus expression vector system, characterized its antiviral potential against NDV, and determined its antiviral potential. Priming of chicken embryo fibroblasts with chIFN-γ elicited an antiviral environment in primary cells, which was mainly due to interferon-stimulated genes (ISGs). A genome-wide transcriptomics approach was used to elucidate the possible signaling pathways associated with IFN-γ-induced immune responses. RNA-sequencing (RNA-seq) data revealed significant induction of ISG-associated pathways, activated temporal expression of ISGs, antiviral mediators, and transcriptional regulators in a cascade of antiviral responses. Collectively, we found that IFN-γ significantly elicited an antiviral response against NDV infection. These data provide a foundation for chIFN-γ-mediated antiviral responses and underpin functional annotation of these important chIFN-γ-induced antiviral influencers.
Keywords: NDV; RNA-seq; chicken embryo fibroblast; interferon; interferon-stimulated gene.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Transcriptome analysis of pre-immune state induced by interferon gamma inhibiting the replication of H9N2 avian influenza viruses in chicken embryo fibroblasts.Infect Genet Evol. 2022 Sep;103:105332. doi: 10.1016/j.meegid.2022.105332. Epub 2022 Jul 8. Infect Genet Evol. 2022. PMID: 35811034
-
Comparative Study of Protection against Newcastle Disease in Young Broilers Administered Natural Chicken Alpha Interferon via Oral and Intramuscular Routes.mSphere. 2020 Jul 15;5(4):e00585-20. doi: 10.1128/mSphere.00585-20. mSphere. 2020. PMID: 32669457 Free PMC article.
-
Expression of interferon gamma by a highly virulent strain of Newcastle disease virus decreases its pathogenicity in chickens.Microb Pathog. 2013 Aug-Sep;61-62:73-83. doi: 10.1016/j.micpath.2013.05.009. Epub 2013 May 25. Microb Pathog. 2013. PMID: 23711962
-
Immune responses of poultry to Newcastle disease virus.Dev Comp Immunol. 2013 Nov;41(3):447-53. doi: 10.1016/j.dci.2013.04.012. Epub 2013 Apr 25. Dev Comp Immunol. 2013. PMID: 23623955 Review.
-
Comprehensive network map of transcriptional activation of chicken type I IFNs and IFN-stimulated genes.Comp Immunol Microbiol Infect Dis. 2020 Feb;68:101407. doi: 10.1016/j.cimid.2019.101407. Epub 2019 Dec 18. Comp Immunol Microbiol Infect Dis. 2020. PMID: 31877494 Review.
Cited by
-
Maternal stevioside supplementation improves intestinal immune function of chicken offspring potentially via modulating gut microbiota and down-regulating the promoter methylation level of suppressor of cytokine signaling 1 (SOCS1).Anim Nutr. 2022 Jun 15;10:329-346. doi: 10.1016/j.aninu.2022.06.002. eCollection 2022 Sep. Anim Nutr. 2022. PMID: 35919247 Free PMC article.
-
Improving the Accumulation and Solubility of Human Interferon-γ (hIFN-γ) in Escherichia coli: A Fusion Protein-Based Method and Network Pharmacology Analysis.Appl Biochem Biotechnol. 2025 Jul 16. doi: 10.1007/s12010-025-05331-z. Online ahead of print. Appl Biochem Biotechnol. 2025. PMID: 40668526
-
Newcastle disease virus suppresses antigen presentation via inhibiting IL-12 expression in dendritic cells.J Zhejiang Univ Sci B. 2024 Mar 15;25(3):254-270. doi: 10.1631/jzus.B2300134. J Zhejiang Univ Sci B. 2024. PMID: 38453639 Free PMC article.
-
In Vitro Characterization of chIFITMs of Aseel and Kadaknath Chicken Breeds against Newcastle Disease Virus Infection.Biology (Basel). 2023 Jun 27;12(7):919. doi: 10.3390/biology12070919. Biology (Basel). 2023. PMID: 37508350 Free PMC article.
-
Antiviral Activity of Porcine IFN-λ3 and IFN-α against Porcine Rotavirus In Vitro.Molecules. 2022 Jul 18;27(14):4575. doi: 10.3390/molecules27144575. Molecules. 2022. PMID: 35889447 Free PMC article.
References
-
- Aldous E, Alexander D. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol 2001, 30: 117–128. - PubMed
-
- Dortmans J, Rottier P, Koch G, Peeters B. Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. J Gen Virol 2011, 92: 336–345. - PubMed
-
- Kommers GD, King DJ, Seal BS, Brown CC. Pathogenesis of chicken-passaged Newcastle disease viruses isolated from chickens and wild and exotic birds. Avian Dis 2003, 47: 319–329. - PubMed
-
- Kommers GD, King DJ, Seal BS, Brown CC. Virulence of six heterogeneous-origin Newcastle disease virus isolates before and after sequential passages in domestic chickens. Avian Pathol 2003, 32: 81–93. - PubMed

