Diagnostic accuracy of genetic markers and nucleic acid techniques for the detection of Leptospira in clinical samples: A meta-analysis
- PMID: 32049960
- PMCID: PMC7041858
- DOI: 10.1371/journal.pntd.0008074
Diagnostic accuracy of genetic markers and nucleic acid techniques for the detection of Leptospira in clinical samples: A meta-analysis
Abstract
Background: Leptospirosis is often difficult to diagnose because of its nonspecific symptoms. The drawbacks of direct isolation and serological tests have led to the increased development of nucleic acid-based assays, which are more rapid and accurate. A meta-analysis was performed to evaluate the diagnostic accuracy of genetic markers for the detection of Leptospira in clinical samples.
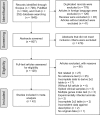
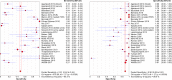
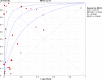
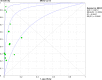
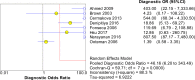
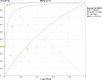
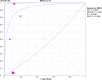
Methodology and principle findings: A literature search was performed in Scopus, PubMed, MEDLINE and non-indexed citations (via Ovid) by using suitable keyword combinations. Studies evaluating the performance of nucleic acid assays targeting leptospire genes in human or animal clinical samples against a reference test were included. Of the 1645 articles identified, 42 eligible studies involving 7414 samples were included in the analysis. The diagnostic performance of nucleic acid assays targeting the rrs, lipL32, secY and flaB genes was pooled and analyzed. Among the genetic markers analyzed, the secY gene showed the highest diagnostic accuracy measures, with a pooled sensitivity of 0.56 (95% CI: 0.50-0.63), a specificity of 0.98 (95% CI: 0.97-0.98), a diagnostic odds ratio of 46.16 (95% CI: 6.20-343.49), and an area under the curve of summary receiver operating characteristics curves of 0.94. Nevertheless, a high degree of heterogeneity was observed in this meta-analysis. Therefore, the present findings here should be interpreted with caution.
Conclusion: The diagnostic accuracies of the studies examined for each genetic marker showed a significant heterogeneity. The secY gene exhibited higher diagnostic accuracy measures compared with other genetic markers, such as lipL32, flaB, and rrs, but the difference was not significant. Thus, these genetic markers had no significant difference in diagnostic accuracy for leptospirosis. Further research into these genetic markers is warranted.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Rapid and sensitive point-of-care detection of Leptospira by RPA-CRISPR/Cas12a targeting lipL32.PLoS Negl Trop Dis. 2022 Jan 6;16(1):e0010112. doi: 10.1371/journal.pntd.0010112. eCollection 2022 Jan. PLoS Negl Trop Dis. 2022. PMID: 34990457 Free PMC article.
-
Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipL32 genes for human leptospirosis in Thailand: a case-control study.PLoS One. 2011 Jan 24;6(1):e16236. doi: 10.1371/journal.pone.0016236. PLoS One. 2011. PMID: 21283633 Free PMC article.
-
Development and validation of a new loop-mediated isothermal amplification for detection of pathogenic Leptospira species in clinical materials.J Microbiol Methods. 2017 Oct;141:55-59. doi: 10.1016/j.mimet.2017.07.010. Epub 2017 Jul 26. J Microbiol Methods. 2017. PMID: 28756184
-
Evaluation of diagnostic accuracy of loop-mediated isothermal amplification method (LAMP) compared with polymerase chain reaction (PCR) for Leptospira spp. in clinical samples: a systematic review and meta-analysis.Diagn Microbiol Infect Dis. 2021 Jul;100(3):115369. doi: 10.1016/j.diagmicrobio.2021.115369. Epub 2021 Mar 13. Diagn Microbiol Infect Dis. 2021. PMID: 33845305
-
Molecular diagnostics for human leptospirosis.Curr Opin Infect Dis. 2016 Oct;29(5):440-5. doi: 10.1097/QCO.0000000000000295. Curr Opin Infect Dis. 2016. PMID: 27537829 Free PMC article. Review.
Cited by
-
Leptospirosis in India: insights on circulating serovars, research lacunae and proposed strategies to control through one health approach.One Health Outlook. 2024 Jun 7;6(1):11. doi: 10.1186/s42522-024-00098-5. One Health Outlook. 2024. PMID: 38849946 Free PMC article. Review.
-
Leptospirosis Cases During the 2024 Catastrophic Flood in Rio Grande Do Sul, Brazil.Pathogens. 2025 Apr 17;14(4):393. doi: 10.3390/pathogens14040393. Pathogens. 2025. PMID: 40333217 Free PMC article.
-
A versatile isothermal amplification assay for the detection of leptospires from various sample types.PeerJ. 2022 Mar 10;10:e12850. doi: 10.7717/peerj.12850. eCollection 2022. PeerJ. 2022. PMID: 35291487 Free PMC article.
-
Genetic Characterization and Zoonotic Potential of Leptospira interrogans Identified in Small Non-Flying Mammals from Southeastern Atlantic Forest, Brazil.Trop Med Infect Dis. 2025 Feb 27;10(3):62. doi: 10.3390/tropicalmed10030062. Trop Med Infect Dis. 2025. PMID: 40137816 Free PMC article.
-
Production and characterization of immunoglobulin G anti-rLipL32 antibody as a biomarker for the diagnosis of leptospirosis.Vet World. 2024 Apr;17(4):871-879. doi: 10.14202/vetworld.2024.871-879. Epub 2024 Apr 19. Vet World. 2024. PMID: 38798296 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

