Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus
- PMID: 32050503
- PMCID: PMC7158669
- DOI: 10.3390/vetsci7010020
Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus
Abstract
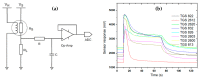
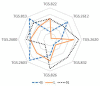
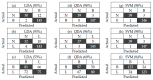
The aim of this study is to determine the performance of a lab-made electronic nose (e-nose) composed of an array of metal oxide semiconductor (MOS) gas sensors in the detection and differentiation of Listeria monocytogenes (L. monocytogenes) and Bacillus cereus (B. cereus) incubated in trypticsoy broth (TSB) media. Conventionally, the detection of L. monocytogenes and B. cereus is often performed by enzyme link immunosorbent assay (ELISA) and polymerase chain reaction (PCR). These techniques require trained operators and expert, expensive reagents and specific containment. In this study, three types of samples, namely, TSB media, L. monocytogenes (serotype 4b American Type Culture Collection (ATCC) 13792), and B. cereus (ATCC) 10876, were used for this experiment. Prior to measurement using the e-nose, each bacterium was inoculated in TSB at 1 × 103-104 CFU/mL, followed by incubation for 48 h. To evaluate the performance of the e-nose, the measured data were then analyzed with chemometric models, namely linear and quadratic discriminant analysis (LDA and QDA), and support vector machine (SVM). As a result, the e-nose coupled with SVM showeda high accuracy of 98% in discriminating between TSB media and L. monocytogenes, and between TSB media and B. cereus. It could be concluded that the lab-made e-nose is able to detect rapidly the presence of bacteria L. monocytogenes and B. cereus on TSB media. For the future, it could be used to identify the presence of L. monocytogenes or B. cereus contamination in the routine and fast assessment of food products in animal quarantine.
Keywords: Bacillus cereus; LDA; Listeria monocytogenes; QDA; SVM; electronic nose.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The inhibitory effect of natural microflora of food on growth of Listeria monocytogenes in enrichment broths.Int J Food Microbiol. 2011 Jan 31;145(1):98-105. doi: 10.1016/j.ijfoodmicro.2010.11.036. Epub 2010 Dec 2. Int J Food Microbiol. 2011. PMID: 21176988
-
Assessment of the microbial safety and quality of cooked chilled foods and their production process.Int J Food Microbiol. 2013 Jan 1;160(3):193-200. doi: 10.1016/j.ijfoodmicro.2012.10.010. Epub 2012 Oct 22. Int J Food Microbiol. 2013. PMID: 23290224
-
A multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus in Korean ready-to-eat food.Foodborne Pathog Dis. 2014 Jul;11(7):574-80. doi: 10.1089/fpd.2013.1638. Epub 2014 May 5. Foodborne Pathog Dis. 2014. PMID: 24796416
-
Development of a random genomic DNA microarray for the detection and identification of Listeria monocytogenes in milk.Int J Food Microbiol. 2013 Feb 1;161(2):134-41. doi: 10.1016/j.ijfoodmicro.2012.11.023. Epub 2012 Dec 6. Int J Food Microbiol. 2013. PMID: 23313851
-
[Comparison of direct colony count methods and the MPN-method for quantitative detection of Listeria in model and field conditions].Berl Munch Tierarztl Wochenschr. 2001 Nov-Dec;114(11-12):453-64. Berl Munch Tierarztl Wochenschr. 2001. PMID: 11766274 Review. German.
Cited by
-
Recent Advances and Applications of Rapid Microbial Assessment from a Food Safety Perspective.Sensors (Basel). 2022 Apr 6;22(7):2800. doi: 10.3390/s22072800. Sensors (Basel). 2022. PMID: 35408414 Free PMC article. Review.
-
2D nanomaterial sensing array using machine learning for differential profiling of pathogenic microbial taxonomic identification.Mikrochim Acta. 2022 Jul 6;189(8):273. doi: 10.1007/s00604-022-05368-5. Mikrochim Acta. 2022. PMID: 35792975 Free PMC article.
-
Recent Advances on Peptide-Based Biosensors and Electronic Noses for Foodborne Pathogen Detection.Biosensors (Basel). 2023 Feb 11;13(2):258. doi: 10.3390/bios13020258. Biosensors (Basel). 2023. PMID: 36832024 Free PMC article. Review.
-
Recording the Presence of Peanibacillus larvae larvae Colonies on MYPGP Substrates Using a Multi-Sensor Array Based on Solid-State Gas Sensors.Sensors (Basel). 2021 Jul 19;21(14):4917. doi: 10.3390/s21144917. Sensors (Basel). 2021. PMID: 34300655 Free PMC article.
-
A Lab-Made E-Nose-MOS Device for Assessing the Bacterial Growth in a Solid Culture Medium.Biosensors (Basel). 2022 Dec 24;13(1):19. doi: 10.3390/bios13010019. Biosensors (Basel). 2022. PMID: 36671854 Free PMC article.
References
-
- Latha C., Sunil B., Kumar V.J., Anu C.J., Deepa J. Evaluation of various cultural enrichment methods for the detection of selected food borne bacterial pathogens. Vet. World. 2014;7:172–176. doi: 10.14202/vetworld.2014.172-176. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

