Physiological and Pathological Roles of RAD52 at DNA Replication Forks
- PMID: 32050645
- PMCID: PMC7072239
- DOI: 10.3390/cancers12020402
Physiological and Pathological Roles of RAD52 at DNA Replication Forks
Abstract
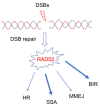
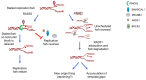
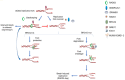
Understanding basic molecular mechanisms underlying the biology of cancer cells is of outmost importance for identification of novel therapeutic targets and biomarkers for patient stratification and better therapy selection. One of these mechanisms, the response to replication stress, fuels cancer genomic instability. It is also an Achille's heel of cancer. Thus, identification of pathways used by the cancer cells to respond to replication-stress may assist in the identification of new biomarkers and discovery of new therapeutic targets. Alternative mechanisms that act at perturbed DNA replication forks and involve fork degradation by nucleases emerged as crucial for sensitivity of cancer cells to chemotherapeutics agents inducing replication stress. Despite its important role in homologous recombination and recombinational repair of DNA double strand breaks in lower eukaryotes, RAD52 protein has been considered dispensable in human cells and the full range of its cellular functions remained unclear. Very recently, however, human RAD52 emerged as an important player in multiple aspects of replication fork metabolism under physiological and pathological conditions. In this review, we describe recent advances on RAD52's key functions at stalled or collapsed DNA replication forks, in particular, the unexpected role of RAD52 as a gatekeeper, which prevents unscheduled processing of DNA. Last, we will discuss how these functions can be exploited using specific inhibitors in targeted therapy or for an informed therapy selection.
Keywords: RAD52; genome stability; replication fork recovery; replication fork reversal; target therapy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

