Amniotic fluid cell-free transcriptome: a glimpse into fetal development and placental cellular dynamics during normal pregnancy
- PMID: 32050959
- PMCID: PMC7017452
- DOI: 10.1186/s12920-020-0690-5
Amniotic fluid cell-free transcriptome: a glimpse into fetal development and placental cellular dynamics during normal pregnancy
Abstract
Background: The amniotic fluid (AF) cell-free transcriptome is modulated by physiologic and pathologic processes during pregnancy. AF gene expression changes with advancing gestation reflect fetal development and organ maturation; yet, defining normal expression and splicing patterns for biomarker discovery in obstetrics requires larger heterogeneous cohorts, evaluation of potential confounding factors, and novel analytical approaches.
Methods: Women with a normal pregnancy who had an AF sample collected during midtrimester (n = 30) or at term gestation (n = 68) were included. Expression profiling at exon level resolution was performed using Human Transcriptome Arrays. Differential expression was based on moderated t-test adjusted p < 0.05 and fold change > 1.25; for differential splicing, a splicing index > 2 and adjusted p < 0.05 were required. Functional profiling was used to interpret differentially expressed or spliced genes. The expression of tissue-specific and cell-type specific signatures defined by single-cell genomics was quantified and correlated with covariates. In-silico validation studies were performed using publicly available datasets.
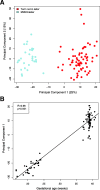
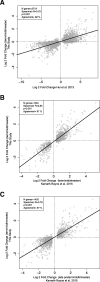
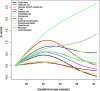
Results: 1) 64,071 genes were detected in AF, with 11% of the coding and 6% of the non-coding genes being differentially expressed between midtrimester and term gestation. Expression changes were highly correlated with those previously reported (R > 0.79, p < 0.001) and featured increased expression of genes specific to the trachea, salivary glands, and lung and decreased expression of genes specific to the cardiac myocytes, uterus, and fetal liver, among others. 2) Single-cell RNA-seq signatures of the cytotrophoblast, Hofbauer cells, erythrocytes, monocytes, T and B cells, among others, showed complex patterns of modulation with gestation (adjusted p < 0.05). 3) In 17% of the genes detected, we found differential splicing with advancing gestation in genes related to brain development processes and immunity pathways, including some that were missed based on differential expression analysis alone.
Conclusions: This represents the largest AF transcriptomics study in normal pregnancy, reporting for the first time that single-cell genomic signatures can be tracked in the AF and display complex patterns of expression during gestation. We also demonstrate a role for alternative splicing in tissue-identity acquisition, organ development, and immune processes. The results herein may have implications for the development of fetal testing to assess placental function and fetal organ maturity.
Keywords: Cell-free RNA; Differential expression; Differential splicing; Fetal sex; Gestational age; Maternal obesity; Single-cell genomic signature; Tissue-specific signature.
Conflict of interest statement
The authors report no conflicts of interest. Dr. Romero has contributed to this work as part of his official duties as an employee of the United States Federal Government.
Figures



References
-
- Cherry SH. Amniotic fluid analysis as an index of fetal health in utero. Med Times. 1967;95(7):713–717. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

