The wild species genome ancestry of domestic chickens
- PMID: 32050971
- PMCID: PMC7014787
- DOI: 10.1186/s12915-020-0738-1
The wild species genome ancestry of domestic chickens
Abstract
Background: Hybridisation and introgression play key roles in the evolutionary history of animal species. They are commonly observed within several orders in wild birds. The domestic chicken Gallus gallus domesticus is the most common livestock species. More than 65 billion chickens are raised annually to produce meat and 80 million metric tons of egg for global human consumption by the commercial sector. Unravelling the origin of its genetic diversity has major application for sustainable breeding improvement programmes.
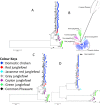
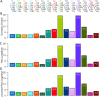
Results: In this study, we report genome-wide analyses for signatures of introgression between indigenous domestic village chicken and the four wild Gallus species. We first assess the genome-wide phylogeny and divergence time across the genus Gallus. Genome-wide sequence divergence analysis supports a sister relationship between the Grey junglefowl G. sonneratii and Ceylon junglefowl G. lafayettii. Both species form a clade that is sister to the Red junglefowl G. gallus, with the Green junglefowl G. varius the most ancient lineage within the genus. We reveal extensive bidirectional introgression between the Grey junglefowl and the domestic chicken and to a much lesser extent with the Ceylon junglefowl. We identify a single case of Green junglefowl introgression. These introgressed regions include genes with biological functions related to development and immune system.
Conclusions: Our study shows that while the Red junglefowl is the main ancestral species, introgressive hybridisation episodes have impacted the genome and contributed to the diversity of the domestic chicken, although likely at different levels across its geographic range.
Keywords: Chicken domestication; Chicken introgression; Divergence time; Evolution; Galliformes; Gallus species; Genetic diversity; Interspecies hybridisation; Livestock species; Speciation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Darwin C. The variation of animals and plants under domestication, vol. 2. 2. United Kingdom: John Murray; 1868.
-
- Liu Y-P, Wu G-S, Yao Y-G, Miao Y-W, Luikart G, Baig M, et al. Multiple maternal origins of chickens: out of the Asian jungles. Mol Phylogen Evol. 2006;38(1):12–19. - PubMed
-
- Delacour J. The pheasant of the world. 2. England: Saiga Publishing Co. Ltd. Surr GU26 GTD; 1977.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

