Immune cell landscaping reveals a protective role for regulatory T cells during kidney injury and fibrosis
- PMID: 32051345
- PMCID: PMC7098794
- DOI: 10.1172/jci.insight.130651
Immune cell landscaping reveals a protective role for regulatory T cells during kidney injury and fibrosis
Abstract
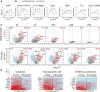
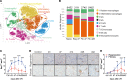
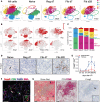
Acute kidney injury (AKI) and chronic kidney diseases are associated with high mortality and morbidity. Although the underlying mechanisms determining the transition from acute to chronic injury are not completely understood, immune-mediated processes are critical in renal injury. We have performed a comparison of 2 mouse models leading to either kidney regeneration or fibrosis. Using global gene expression profiling we could identify immune-related pathways accounting for the majority of the observed transcriptional changes during fibrosis. Unbiased examination of the immune cell composition, using single-cell RNA sequencing, revealed major changes in tissue-resident macrophages and T cells. Following injury, there was a marked increase in tissue-resident IL-33R+ and IL-2Ra+ regulatory T cells (Tregs). Expansion of this population before injury protected the kidney from injury and fibrosis. Transcriptional profiling of Tregs showed a differential upregulation of regenerative and proangiogenic pathways during regeneration, whereas in the fibrotic environment they expressed markers of hyperactivation and fibrosis. Our data point to a hitherto underappreciated plasticity in Treg function within the same tissue, dictated by environmental cues. Overall, we provide a detailed cellular and molecular characterization of the immunological changes during kidney injury, regeneration, and fibrosis.
Keywords: Adaptive immunity; Chronic kidney disease; Fibrosis; Immunology; Nephrology.
Conflict of interest statement
Figures


References
-
- Hoste EA, Schurgers M. Epidemiology of acute kidney injury: how big is the problem? Crit Care Med. 2008;36(4 suppl):S146–S151. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

