Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plants under differential biotic stresses
- PMID: 32051495
- PMCID: PMC7015943
- DOI: 10.1038/s41598-020-59168-z
Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plants under differential biotic stresses
Abstract
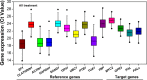
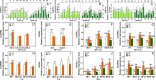
The selection of reliable reference genes (RGs) for normalization under given experimental conditions is necessary to develop an accurate qRT-PCR assay. To the best of our knowledge, only a small number of RGs have been rigorously identified and used in tea plants (Camellia sinensis (L.) O. Kuntze) under abiotic stresses, but no critical RG identification has been performed for tea plants under any biotic stresses till now. In the present study, we measured the mRNA transcriptional levels of ten candidate RGs under five experimental conditions; these genes have been identified as stable RGs in tea plants. By using the ΔCt method, geNorm, NormFinder and BestKeeper, CLATHRIN1 and UBC1, TUA1 and SAND1, or SAND1 and UBC1 were identified as the best combination for normalizing diurnal gene expression in leaves, stems and roots individually; CLATHRIN1 and GAPDH1 were identified as the best combination for jasmonic acid treatment; ACTIN1 and UBC1 were identified as the best combination for Toxoptera aurantii-infested leaves; UBC1 and GAPDH1 were identified as the best combination for Empoasca onukii-infested leaves; and SAND1 and TBP1 were identified as the best combination for Ectropis obliqua regurgitant-treated leaves. Furthermore, our results suggest that if the processing time of the treatment was long, the best RGs for normalization should be recommended according to the stability of the proposed RGs in different time intervals when intragroup differences were compared, which would strongly increase the accuracy and sensitivity of target gene expression in tea plants under biotic stresses. However, when the differences of intergroup were compared, the RGs for normalization should keep consistent across different time points. The results of this study provide a technical guidance for further study of the molecular mechanisms of tea plants under different biotic stresses.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

