Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids
- PMID: 32051547
- PMCID: PMC7423603
- DOI: 10.1038/s41380-020-0676-x
Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids
Abstract
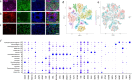
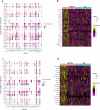
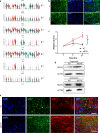
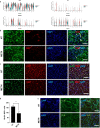
Methamphetamine (METH) is a potent stimulant that induces a euphoric state but also causes cognitive impairment, neurotoxicity and neurodevelopmental deficits. Yet, the molecular mechanisms by which METH causes neurodevelopmental defects have remained elusive. Here we utilized human cerebral organoids and single-cell RNA sequencing (scRNA-seq) to study the effects of prenatal METH exposure on fetal brain development. We analyzed 20,758 cells from eight untreated and six METH-treated cerebral organoids and found that the organoids developed from embryonic stem cells contained a diverse array of glial and neuronal cell types. We further identified transcriptionally distinct populations of astrocytes and oligodendrocytes within cerebral organoids. Treatment of organoids with METH-induced marked changes in transcription in multiple cell types, including astrocytes and neural progenitor cells. METH also elicited novel astrocyte-specific gene expression networks regulating responses to cytokines, and inflammasome. Moreover, upregulation of immediate early genes, complement factors, apoptosis, and immune response genes suggests a neuroinflammatory program induced by METH regulating neural stem cell proliferation, differentiation, and cell death. Finally, we observed marked METH-induced changes in neuroinflammatory and cytokine gene expression at the RNA and protein levels. Our data suggest that human cerebral organoids represent a model system to study drug-induced neuroinflammation at single-cell resolution.
Conflict of interest statement
TMR is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Figures

References
-
- Prakash MD, Tangalakis K, Antonipillai J, Stojanovska L, Nurgali K, Apostolopoulos V. Methamphetamine: effects on the brain, gut and immune system. Pharmacol Res. 2017;120:60–7. - PubMed
-
- Chang L, Alicata D, Ernst T, Volkow N. Structural and metabolic brain changes in the striatum associated with methamphetamine abuse. Addiction. 2007;102(Suppl 1):16–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

