The global Microcystis interactome
- PMID: 32051648
- PMCID: PMC7003799
- DOI: 10.1002/lno.11361
The global Microcystis interactome
Abstract
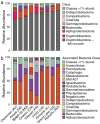
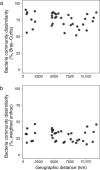
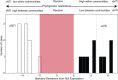
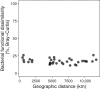
Bacteria play key roles in the function and diversity of aquatic systems, but aside from study of specific bloom systems, little is known about the diversity or biogeography of bacteria associated with harmful cyanobacterial blooms (cyanoHABs). CyanoHAB species are known to shape bacterial community composition and to rely on functions provided by the associated bacteria, leading to the hypothesized cyanoHAB interactome, a coevolved community of synergistic and interacting bacteria species, each necessary for the success of the others. Here, we surveyed the microbiome associated with Microcystis aeruginosa during blooms in 12 lakes spanning four continents as an initial test of the hypothesized Microcystis interactome. We predicted that microbiome composition and functional potential would be similar across blooms globally. Our results, as revealed by 16S rRNA sequence similarity, indicate that M. aeruginosa is cosmopolitan in lakes across a 280° longitudinal and 90° latitudinal gradient. The microbiome communities were represented by a wide range of operational taxonomic units and relative abundances. Highly abundant taxa were more related and shared across most sites and did not vary with geographic distance, thus, like Microcystis, revealing no evidence for dispersal limitation. High phylogenetic relatedness, both within and across lakes, indicates that microbiome bacteria with similar functional potential were associated with all blooms. While Microcystis and the microbiome bacteria shared many genes, whole-community metagenomic analysis revealed a suite of biochemical pathways that could be considered complementary. Our results demonstrate a high degree of similarity across global Microcystis blooms, thereby providing initial support for the hypothesized Microcystis interactome.
© 2019 The Authors. Limnology and Oceanography published by Wiley Periodicals, Inc. on behalf of Association for the Sciences of Limnology and Oceanography.
Conflict of interest statement
None declared.
Figures


References
-
- Batista, A. , Woodhouse J., Grossart H.‐P., and Giani A.. 2018. Methanogenic archaea associated to Microcystis sp. in field samples and in culture. Hydrobiologia 831: 1–10. doi: 10.1007/s10750-018-3655-3 - DOI
-
- Bell, W. , and Mitchell R.. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Bio. Bull. 143: 265–277. doi: 10.2307/1540052 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
