Physiological functions of SPP/SPPL intramembrane proteases
- PMID: 32052089
- PMCID: PMC7366577
- DOI: 10.1007/s00018-020-03470-6
Physiological functions of SPP/SPPL intramembrane proteases
Abstract
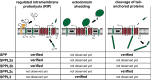
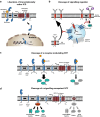
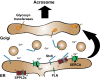
Intramembrane proteolysis describes the cleavage of substrate proteins within their hydrophobic transmembrane segments. Several families of intramembrane proteases have been identified including the aspartyl proteases Signal peptide peptidase (SPP) and its homologues, the SPP-like (SPPL) proteases SPPL2a, SPPL2b, SPPL2c and SPPL3. As presenilin homologues, they employ a similar catalytic mechanism as the well-studied γ-secretase. However, SPP/SPPL proteases cleave transmembrane proteins with a type II topology. The characterisation of SPP/SPPL-deficient mouse models has highlighted a still growing spectrum of biological functions and also promoted the substrate discovery of these proteases. In this review, we will summarise the current hypotheses how phenotypes of these mouse models are linked to the molecular function of the enzymes. At the cellular level, SPP/SPPL-mediated cleavage events rather provide specific regulatory switches than unspecific bulk proteolysis. By this means, a plethora of different cell biological pathways is influenced including signal transduction, membrane trafficking and protein glycosylation.
Keywords: Intramembrane proteolysis; Membrane trafficking; Protein degradation; Signal peptide peptidase-like; Signal transduction; γ-secretase.
Figures

Similar articles
-
The role of SPP/SPPL intramembrane proteases in membrane protein homeostasis.FEBS J. 2024 Jan;291(1):25-44. doi: 10.1111/febs.16941. Epub 2023 Sep 5. FEBS J. 2024. PMID: 37625440 Review.
-
Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b.J Biol Chem. 2008 Jan 18;283(3):1644-1652. doi: 10.1074/jbc.M706661200. Epub 2007 Oct 25. J Biol Chem. 2008. PMID: 17965014
-
Signal peptide peptidase-like 2 proteases: Regulatory switches or proteasome of the membrane?Biochim Biophys Acta Mol Cell Res. 2022 Jan;1869(1):119163. doi: 10.1016/j.bbamcr.2021.119163. Epub 2021 Oct 18. Biochim Biophys Acta Mol Cell Res. 2022. PMID: 34673079 Review.
-
Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases.Biochim Biophys Acta. 2013 Dec;1828(12):2828-39. doi: 10.1016/j.bbamem.2013.03.033. Biochim Biophys Acta. 2013. PMID: 24099004 Review.
-
Signal peptide peptidase and SPP-like proteases - Possible therapeutic targets?Biochim Biophys Acta Mol Cell Res. 2017 Nov;1864(11 Pt B):2169-2182. doi: 10.1016/j.bbamcr.2017.06.007. Epub 2017 Jun 15. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28624439 Review.
Cited by
-
Pan-cancer analysis suggests histocompatibility minor 13 is an unfavorable prognostic biomarker promoting cell proliferation, migration, and invasion in hepatocellular carcinoma.Front Pharmacol. 2022 Aug 15;13:950156. doi: 10.3389/fphar.2022.950156. eCollection 2022. Front Pharmacol. 2022. PMID: 36046831 Free PMC article.
-
Signaling Functions of Intramembrane Aspartyl-Proteases.Front Cardiovasc Med. 2020 Dec 14;7:591787. doi: 10.3389/fcvm.2020.591787. eCollection 2020. Front Cardiovasc Med. 2020. PMID: 33381526 Free PMC article. Review.
-
More Than Just Simple Interaction between STIM and Orai Proteins: CRAC Channel Function Enabled by a Network of Interactions with Regulatory Proteins.Int J Mol Sci. 2021 Jan 5;22(1):471. doi: 10.3390/ijms22010471. Int J Mol Sci. 2021. PMID: 33466526 Free PMC article. Review.
-
Microglial ER stress response via IRE1α regulates diet-induced metabolic imbalance and obesity in mice.Mol Metab. 2025 May;95:102128. doi: 10.1016/j.molmet.2025.102128. Epub 2025 Mar 20. Mol Metab. 2025. PMID: 40120978 Free PMC article.
-
Proteolytic Regulation of the Lectin-Like Oxidized Lipoprotein Receptor LOX-1.Front Cardiovasc Med. 2021 Jan 20;7:594441. doi: 10.3389/fcvm.2020.594441. eCollection 2020. Front Cardiovasc Med. 2021. PMID: 33553253 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

