Sparse identification of contrast gain control in the fruit fly photoreceptor and amacrine cell layer
- PMID: 32052209
- PMCID: PMC7016054
- DOI: 10.1186/s13408-020-0080-5
Sparse identification of contrast gain control in the fruit fly photoreceptor and amacrine cell layer
Abstract
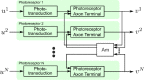
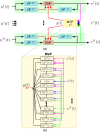
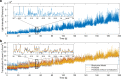
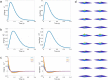
The fruit fly's natural visual environment is often characterized by light intensities ranging across several orders of magnitude and by rapidly varying contrast across space and time. Fruit fly photoreceptors robustly transduce and, in conjunction with amacrine cells, process visual scenes and provide the resulting signal to downstream targets. Here, we model the first step of visual processing in the photoreceptor-amacrine cell layer. We propose a novel divisive normalization processor (DNP) for modeling the computation taking place in the photoreceptor-amacrine cell layer. The DNP explicitly models the photoreceptor feedforward and temporal feedback processing paths and the spatio-temporal feedback path of the amacrine cells. We then formally characterize the contrast gain control of the DNP and provide sparse identification algorithms that can efficiently identify each the feedforward and feedback DNP components. The algorithms presented here are the first demonstration of tractable and robust identification of the components of a divisive normalization processor. The sparse identification algorithms can be readily employed in experimental settings, and their effectiveness is demonstrated with several examples.
Keywords: Contrast gain control; Divisive normalization; Fruit fly; Photoreceptor; Sparse functional identification.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








Similar articles
-
Divisive normalization processors in the early visual system of the Drosophila brain.Biol Cybern. 2023 Dec;117(6):411-431. doi: 10.1007/s00422-023-00972-x. Epub 2023 Sep 13. Biol Cybern. 2023. PMID: 37702831 Free PMC article.
-
Sign-conserving amacrine neurons in the fly's external plexiform layer.Vis Neurosci. 2005 May-Jun;22(3):345-58. doi: 10.1017/S095252380522309X. Vis Neurosci. 2005. PMID: 16079009
-
Ih channels control feedback regulation from amacrine cells to photoreceptors.PLoS Biol. 2015 Apr 1;13(4):e1002115. doi: 10.1371/journal.pbio.1002115. eCollection 2015 Apr. PLoS Biol. 2015. PMID: 25831426 Free PMC article.
-
A retinal dark-light switch: a review of the evidence.Vis Neurosci. 1996 May-Jun;13(3):399-409. doi: 10.1017/s0952523800008087. Vis Neurosci. 1996. PMID: 8782368 Review.
-
Inhibitory Interneurons in the Retina: Types, Circuitry, and Function.Annu Rev Vis Sci. 2017 Sep 15;3:1-24. doi: 10.1146/annurev-vision-102016-061345. Epub 2017 Jun 15. Annu Rev Vis Sci. 2017. PMID: 28617659 Review.
Cited by
-
The functional logic of odor information processing in the Drosophila antennal lobe.PLoS Comput Biol. 2023 Apr 21;19(4):e1011043. doi: 10.1371/journal.pcbi.1011043. eCollection 2023 Apr. PLoS Comput Biol. 2023. PMID: 37083547 Free PMC article.
-
Divisive normalization processors in the early visual system of the Drosophila brain.Biol Cybern. 2023 Dec;117(6):411-431. doi: 10.1007/s00422-023-00972-x. Epub 2023 Sep 13. Biol Cybern. 2023. PMID: 37702831 Free PMC article.
-
Modeling and characterization of pure and odorant mixture processing in the Drosophila mushroom body calyx.Front Physiol. 2024 Oct 16;15:1410946. doi: 10.3389/fphys.2024.1410946. eCollection 2024. Front Physiol. 2024. PMID: 39479309 Free PMC article.
-
Accelerating with FlyBrainLab the discovery of the functional logic of the Drosophila brain in the connectomic and synaptomic era.Elife. 2021 Feb 22;10:e62362. doi: 10.7554/eLife.62362. Elife. 2021. PMID: 33616035 Free PMC article.
References
-
- Laughlin SB. Matching coding, circuits, cells, and molecules to signals: general principles of retinal design in the fly’s eye. Prog Retin Eye Res. 1994;13(1):165–196. doi: 10.1016/1350-9462(94)90009-4. - DOI
-
- Rodieck RW. The first steps in seeing. London: Oxford University Press; 1998.
Grants and funding
LinkOut - more resources
Full Text Sources

