Long noncoding RNA LINC00963 promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1
- PMID: 32052688
- PMCID: PMC7100992
- DOI: 10.1080/15384101.2020.1728024
Long noncoding RNA LINC00963 promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1
Abstract
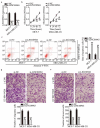
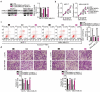
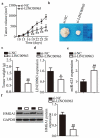
Extensive research has shown that LINC00963 is aberrantly expressed in human cancers, and that dysregulation of LINC00963 is implicated in the initiation and progression of human cancers. The expression and functions of LINC00963 in breast cancer are still unclear. Our aims were to measure the expression of LINC00963 in breast cancer, determine its effects on malignant behaviors of tumor cells, and uncover the molecular events underlying the actions of LINC00963 in breast cancer. Herein, LINC00963 was found to be overexpressed in breast cancer samples, and its overexpression was correlated with lymph node metastasis, TNM stage and differentiation grade. Patients with breast cancer harboring higher LINC00963 expression showed shorter overall survival than did the patients with lower LINC00963 expression. Functional experiments revealed that depletion of LINC00963 inhibited breast cancer cell proliferation, migration, and invasion and facilitated apoptosis in vitro and impaired tumor growth in vivo. Mechanism investigation revealed that LINC00963 can interact with microRNA-625 (miR-625). LINC00963 worked as a competitive endogenous RNA for miR-625 to weaken the suppressive effect of miR-625 on high mobility group AT-hook 1 (HMGA1) in breast cancer cells. Furthermore, miR-625 inhibition and HMGA1 restoration both abrogated the effects of LINC00963 silencing on breast cancer cells. Our findings indicate that the LINC00963-miR-625-HMGA1 pathway plays an important role in the malignancy of breast cancer in vitro and in vivo. Hence, targeting this pathway may be a novel strategy against breast cancer.
Keywords: High mobility group AT-hook 1; LINC00963; breast cancer; microRNA-625.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
