Comparison of paired cerebrospinal fluid and serum cell-free mitochondrial and nuclear DNA with copy number and fragment length
- PMID: 32052892
- PMCID: PMC7307366
- DOI: 10.1002/jcla.23238
Comparison of paired cerebrospinal fluid and serum cell-free mitochondrial and nuclear DNA with copy number and fragment length
Abstract
Background: Most studies on cell-free DNA (cfDNA) were only for single body fluids; however, the differences in cfDNA distribution between two body fluids are rarely reported. Hence, in this work, we compared the differences in cfDNA distribution between cerebrospinal fluid (CSF) and serum of patients with brain-related diseases.
Methods: The fragment length of cfDNA was determined by using Agilent 2100 Bioanalyzer. The copy numbers of cell-free mitochondrial DNA (cf-mtDNA) and cell-free nuclear DNA (cf-nDNA) were determined by using real-time quantitative PCR (qPCR) and droplet digital PCR (ddPCR) with three pairs of mitochondrial ND1 and nuclear GAPDH primers, respectively.
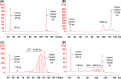
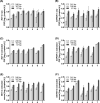
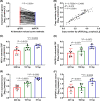
Results: There were short (~60 bp), medium (~167 bp), and long (>250 bp) cfDNA fragment length distributions totally obtained from CSF and serum using Agilent 2100 Bioanalyzer. The results of both qPCR and ddPCR confirmed the existence of these three cfDNA fragment ranges in CSF and serum. According to qPCR, the copy numbers of long cf-mtDNA, medium, and long cf-nDNA in CSF were significantly higher than in paired serum. In CSF, only long cf-mtDNA's copy numbers were higher than long cf-nDNA. But in serum, the copy numbers of medium and long cf-mtDNA were higher than the corresponding cf-nDNA.
Conclusion: The cf-nDNA and cf-mtDNA with different fragment lengths differentially distributed in the CSF and serum of patients with brain disorders, which might serve as a biomarker of human brain diseases.
Keywords: cell-free mitochondrial DNA; cell-free nuclear DNA; cerebrospinal fluid; copy number; fragment length; serum.
© 2020 The Authors. Journal of Clinical Laboratory Analysis published by Wiley Periodicals, Inc.
Conflict of interest statement
All authors in this study had no conflicts of interest.
Figures



Similar articles
-
Analysis of cf-mtDNA and cf-nDNA fragment size distribution using different isolation methods in BV-2 cell supernatant of starvation-induced autophagy.Biochim Biophys Acta Mol Cell Res. 2022 Jan;1869(1):119147. doi: 10.1016/j.bbamcr.2021.119147. Epub 2021 Sep 30. Biochim Biophys Acta Mol Cell Res. 2022. PMID: 34600918
-
Mitochondrial dysfunction in cumulus-oocyte complexes increases cell-free mitochondrial DNA.J Reprod Dev. 2018 Jun 22;64(3):261-266. doi: 10.1262/jrd.2018-012. Epub 2018 Apr 3. J Reprod Dev. 2018. PMID: 29618676 Free PMC article.
-
Probing the diagnostic values of plasma cf-nDNA and cf-mtDNA for Parkinson's disease and multiple system atrophy.Front Neurosci. 2024 Dec 2;18:1488820. doi: 10.3389/fnins.2024.1488820. eCollection 2024. Front Neurosci. 2024. PMID: 39687490 Free PMC article.
-
Absolute Quantification of Cellular and Cell-Free Mitochondrial DNA Copy Number from Human Blood and Urinary Samples Using Real Time Quantitative PCR.Methods Mol Biol. 2025;2878:233-257. doi: 10.1007/978-1-0716-4264-1_13. Methods Mol Biol. 2025. PMID: 39546266
-
Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application.Int J Mol Sci. 2020 Sep 17;21(18):6827. doi: 10.3390/ijms21186827. Int J Mol Sci. 2020. PMID: 32957662 Free PMC article. Review.
Cited by
-
The Value of Whole-Genome Sequencing for Mitochondrial DNA Population Studies: Strategies and Criteria for Extracting High-Quality Mitogenome Haplotypes.Int J Mol Sci. 2022 Feb 17;23(4):2244. doi: 10.3390/ijms23042244. Int J Mol Sci. 2022. PMID: 35216360 Free PMC article.
-
Clinical Circulating Tumor DNA Testing for Precision Oncology.Cancer Res Treat. 2023 Apr;55(2):351-366. doi: 10.4143/crt.2022.1026. Cancer Res Treat. 2023. PMID: 36915242 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

