Context-Specific Nested Effects Models
- PMID: 32053004
- PMCID: PMC7081248
- DOI: 10.1089/cmb.2019.0459
Context-Specific Nested Effects Models
Abstract
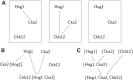
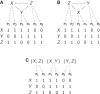
Advances in systems biology have made clear the importance of network models for capturing knowledge about complex relationships in gene regulation, metabolism, and cellular signaling. A common approach to uncovering biological networks involves performing perturbations on elements of the network, such as gene knockdown experiments, and measuring how the perturbation affects some reporter of the process under study. In this article, we develop context-specific nested effects models (CSNEMs), an approach to inferring such networks that generalizes nested effects models (NEMs). The main contribution of this work is that CSNEMs explicitly model the participation of a gene in multiple contexts, meaning that a gene can appear in multiple places in the network. Biologically, the representation of regulators in multiple contexts may indicate that these regulators have distinct roles in different cellular compartments or cell cycle phases. We present an evaluation of the method on simulated data as well as on data from a study of the sodium chloride stress response in Saccharomyces cerevisiae.
Keywords: context specific; graph; inference; nested effects models; network.
Conflict of interest statement
The authors declare they have no competing financial interests.
Figures






References
-
- Friedman N., Linial M., Nachman I., et al. . 2000. Using Bayesian networks to analyze expression data. J. Comput. Biol. 7, 601–620 - PubMed
-
- Froehlich H., Markowetz F., Tresch A., et al. nem: (Dynamic) Nested Effects Models and Deterministic Effects Propagation Networks to Reconstruct Phenotypic Hierarchies. R Package Version 2.60.0 2019
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

