A rapid and simple bead-bashing-based method for genomic DNA extraction from mammalian tissue
- PMID: 32054310
- PMCID: PMC7252492
- DOI: 10.2144/btn-2019-0172
A rapid and simple bead-bashing-based method for genomic DNA extraction from mammalian tissue
Abstract
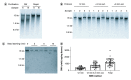
Conventional genomic DNA (gDNA) extraction methods can take hours to complete, may require fume hoods and represent the most time-consuming step in many gDNA-based molecular assays. We systematically optimized a bead bashing-based (BBB) approach for rapid gDNA extraction without the need for a fume hood. Human tissue specimens (n = 34) subjected to the 12-min BBB method yielded 0.40 ± 0.17 (mean ± SD) μg of gDNA per milligram of tissue, sufficient for many downstream applications, and 3- and 6-min extensions resulted in an additional 0.43 ± 0.23 μg and 0.48 ± 0.43 μg per milligram of tissue, respectively. The BBB method provides a simple and rapid method for gDNA extraction from mammalian tissue that is applicable to time-sensitive clinical applications.
Keywords: genetic testing; rapid clinical diagnostics; rapid tissue gDNA extraction; tissue processing.
Conflict of interest statement
S Wei and Z Williams are listed as inventors on patents relating to this technology filed by Columbia University. This research was supported by the National Institutes of Health grants HD068546, HD100013 and U19CA179564, and Columbia BioMedX. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures

Similar articles
-
Development of a Membrane-Based Method for Isolation of Genomic DNA from Human Blood.J Biomol Tech. 2018 Jul;29(2):46-53. doi: 10.7171/jbt.18-2902-001. Epub 2018 Mar 23. J Biomol Tech. 2018. PMID: 29623006 Free PMC article.
-
Efficiency comparison of three methods for extracting genomic DNA of the pathogenic oomycete Pythium insidiosum.J Med Assoc Thai. 2014 Mar;97(3):342-8. J Med Assoc Thai. 2014. PMID: 25123015
-
Automated and robust extraction of genomic DNA from various leftover blood samples.Anal Biochem. 2023 Oct 1;678:115271. doi: 10.1016/j.ab.2023.115271. Epub 2023 Aug 3. Anal Biochem. 2023. PMID: 37543277
-
Isolation of human genomic DNA for genetic analysis from premature neonates: a comparison between newborn dried blood spots, whole blood and umbilical cord tissue.BMC Genet. 2013 Oct 29;14:105. doi: 10.1186/1471-2156-14-105. BMC Genet. 2013. PMID: 24168095 Free PMC article.
-
An innovative optimized protocol for high-quality genomic DNA extraction from recalcitrant Shea tree (Vitellaria paradoxa, C.F. Gaertn) plant and its suitability for downstream applications.Mol Biol Rep. 2024 Jan 22;51(1):171. doi: 10.1007/s11033-023-09098-6. Mol Biol Rep. 2024. PMID: 38252378
Cited by
-
DNA Content of Various Fluids and Tissues of the Human Body.Genes (Basel). 2023 Dec 21;15(1):17. doi: 10.3390/genes15010017. Genes (Basel). 2023. PMID: 38275599 Free PMC article.
-
Nuclei on the Rise: When Nuclei-Based Methods Meet Next-Generation Sequencing.Cells. 2023 Mar 30;12(7):1051. doi: 10.3390/cells12071051. Cells. 2023. PMID: 37048124 Free PMC article. Review.
-
Development and validation of a flexible DNA extraction (PAN) method for liquid biopsy of multiple sample types.J Clin Lab Anal. 2021 Sep;35(9):e23962. doi: 10.1002/jcla.23962. Epub 2021 Aug 16. J Clin Lab Anal. 2021. PMID: 34399000 Free PMC article.
-
Molecular characterization of chlorpyrifos degrading bacteria isolated from contaminated dairy farm soils in Nakuru County, Kenya.Heliyon. 2022 Mar 24;8(3):e09176. doi: 10.1016/j.heliyon.2022.e09176. eCollection 2022 Mar. Heliyon. 2022. PMID: 35846483 Free PMC article.
References
-
- Zhang J, Walsh MF, Wu G. et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373(24), 2336–2346 (2015). - PMC - PubMed
-
• A simple, rapid and robust gDNA extraction method from mammalian tissue can be applied on and can significantly speed up the the listed studies.
-
- Carp H, Guetta E, Dorf H, Soriano D, Barkai G, Schiff E. Embryonic karyotype in recurrent miscarriage with parental karyotypic aberrations. Fertil. Steril. 85(2), 446–450 (2006). - PubMed
-
- De Paoli-Iseppi R, Johansson PA, Menzies AM. et al. Comparison of whole-exome sequencing of matched fresh and formalin fixed paraffin embedded melanoma tumours: implications for clinical decision making. Pathology 48(3), 261–266 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
