Natural images are reliably represented by sparse and variable populations of neurons in visual cortex
- PMID: 32054847
- PMCID: PMC7018721
- DOI: 10.1038/s41467-020-14645-x
Natural images are reliably represented by sparse and variable populations of neurons in visual cortex
Abstract
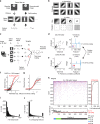
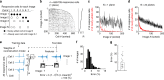
Natural scenes sparsely activate neurons in the primary visual cortex (V1). However, how sparsely active neurons reliably represent complex natural images and how the information is optimally decoded from these representations have not been revealed. Using two-photon calcium imaging, we recorded visual responses to natural images from several hundred V1 neurons and reconstructed the images from neural activity in anesthetized and awake mice. A single natural image is linearly decodable from a surprisingly small number of highly responsive neurons, and the remaining neurons even degrade the decoding. Furthermore, these neurons reliably represent the image across trials, regardless of trial-to-trial response variability. Based on our results, diverse, partially overlapping receptive fields ensure sparse and reliable representation. We suggest that information is reliably represented while the corresponding neuronal patterns change across trials and collecting only the activity of highly responsive neurons is an optimal decoding strategy for the downstream neurons.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

