Inference and effects of barcode multiplets in droplet-based single-cell assays
- PMID: 32054859
- PMCID: PMC7018801
- DOI: 10.1038/s41467-020-14667-5
Inference and effects of barcode multiplets in droplet-based single-cell assays
Abstract
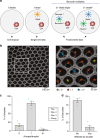
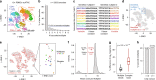
A widespread assumption for single-cell analyses specifies that one cell's nucleic acids are predominantly captured by one oligonucleotide barcode. Here, we show that ~13-21% of cell barcodes from the 10x Chromium scATAC-seq assay may have been derived from a droplet with more than one oligonucleotide sequence, which we call "barcode multiplets". We demonstrate that barcode multiplets can be derived from at least two different sources. First, we confirm that approximately 4% of droplets from the 10x platform may contain multiple beads. Additionally, we find that approximately 5% of beads may contain detectable levels of multiple oligonucleotide barcodes. We show that this artifact can confound single-cell analyses, including the interpretation of clonal diversity and proliferation of intra-tumor lymphocytes. Overall, our work provides a conceptual and computational framework to identify and assess the impacts of barcode multiplets in single-cell data.
Conflict of interest statement
The authors declare the following competing interests: J.D.B. holds patents related to ATAC-seq. All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

