Modeling membrane nanotube morphology: the role of heterogeneity in composition and material properties
- PMID: 32054874
- PMCID: PMC7018976
- DOI: 10.1038/s41598-020-59221-x
Modeling membrane nanotube morphology: the role of heterogeneity in composition and material properties
Abstract
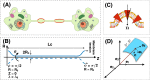
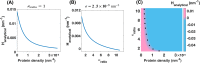
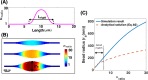
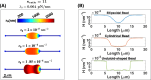
Membrane nanotubes are dynamic structures that may connect cells over long distances. Nanotubes are typically thin cylindrical tubes, but they may occasionally have a beaded architecture along the tube. In this paper, we study the role of membrane mechanics in governing the architecture of these tubes and show that the formation of bead-like structures along the nanotubes can result from local heterogeneities in the membrane either due to protein aggregation or due to membrane composition. We present numerical results that predict how membrane properties, protein density, and local tension compete to create a phase space that governs the morphology of a nanotube. We also find that there exists a discontinuity in the energy that impedes two beads from fusing. These results suggest that the membrane-protein interaction, membrane composition, and membrane tension closely govern the tube radius, number of beads, and the bead morphology.
Conflict of interest statement
The authors declare no competing interests.
Figures