An atypical short-chain dehydrogenase-reductase functions in the relaxation of photoprotective qH in Arabidopsis
- PMID: 32055052
- PMCID: PMC7288749
- DOI: 10.1038/s41477-020-0591-9
An atypical short-chain dehydrogenase-reductase functions in the relaxation of photoprotective qH in Arabidopsis
Abstract
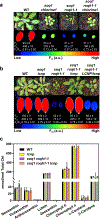
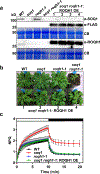
Photosynthetic organisms experience wide fluctuations in light intensity and regulate light harvesting accordingly to prevent damage from excess energy. The antenna quenching component qH is a sustained form of energy dissipation that protects the photosynthetic apparatus under stress conditions. This photoprotective mechanism requires the plastid lipocalin LCNP and is prevented by SUPPRESSOR OF QUENCHING1 (SOQ1) under non-stress conditions. However, the molecular mechanism of qH relaxation has yet to be resolved. Here, we isolated and characterized RELAXATION OF QH1 (ROQH1), an atypical short-chain dehydrogenase-reductase that functions as a qH-relaxation factor in Arabidopsis. The ROQH1 gene belongs to the GreenCut2 inventory specific to photosynthetic organisms, and the ROQH1 protein localizes to the chloroplast stroma lamellae membrane. After a cold and high-light treatment, qH does not relax in roqh1 mutants and qH does not occur in leaves overexpressing ROQH1. When the soq1 and roqh1 mutations are combined, qH can neither be prevented nor relaxed and soq1 roqh1 displays constitutive qH and light-limited growth. We propose that LCNP and ROQH1 perform dosage-dependent, antagonistic functions to protect the photosynthetic apparatus and maintain light-harvesting efficiency in plants.
Conflict of interest statement
Competing interest statement
The authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures









References
-
- Niyogi KK Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol 50, 333–359 (1999). - PubMed
-
- Demmig-Adams B & Adams WW III Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Phvsiol. Plant Mol. Bioi 43, 599–626 (1992).
-
- Walters RG & Horton P Resolution of components of non-photochemical chlorophyll fluorescence quenching in barley leaves. Photosynth. Res 27, 121–133 (1991). - PubMed
-
- Malnoë A Photoinhibition or photoprotection of photosynthesis, which one is it? Update on newly termed sustained quenching component, qH. Environ. Exp. Bot (2018). doi: 10.1016/j.envexpbot.2018.05.005 - DOI
-
- Li X-P et al. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem 279, 22866–22874 (2004). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

